7ENT
 
 | |
7EV2
 
 | |
6KEE
 
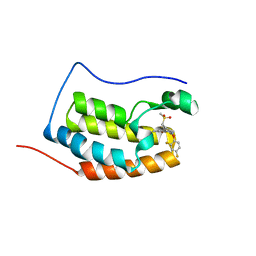 | | Crystal structure of BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12154651 Å) | | Cite: | Crystal structure of BRD4 Bromodomain1 with an inhibitor
To be published
|
|
6KEG
 
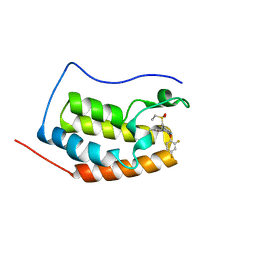 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(ethylsulfonylmethyl)pyridin-3-yl]-8-methyl-4H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KEF
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(8-methyl-1-oxidanylidene-2H-pyrrolo[1,2-a]pyrazin-6-yl)-4-phenoxy-phenyl]methanesulfonamide | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44467545 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KED
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-d][1,2,4]triazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.548447 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
7ELT
 
 | |
6IV3
 
 | |
6IV0
 
 | |
6IV2
 
 | |
6JLF
 
 | |
6IV1
 
 | |
7WOT
 
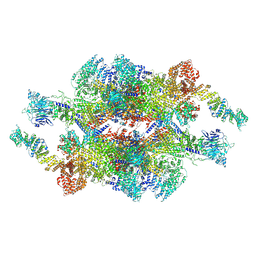 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
5XUX
 
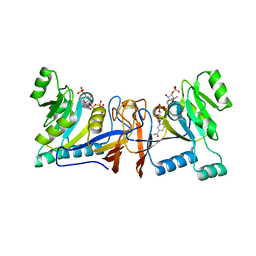 | | Crystal structure of Rib7 from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV0
 
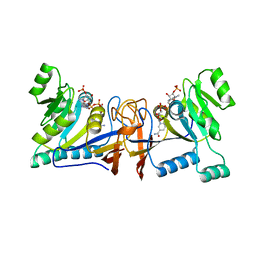 | | Crystal structure of Rib7 mutant D33N from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV2
 
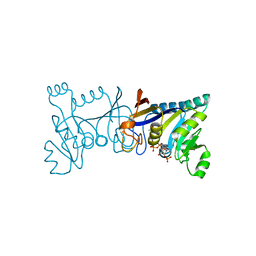 | | Crystal structure of Rib7 mutant D33A from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7WOO
 
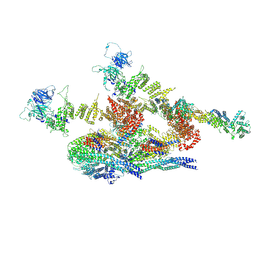 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
5XV5
 
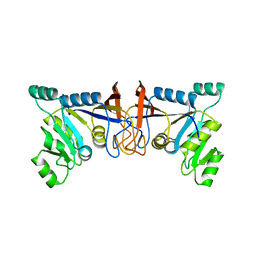 | | Crystal structure of Rib7 mutant S88E from Methanosarcina mazei | | Descriptor: | Conserved protein | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6IVW
 
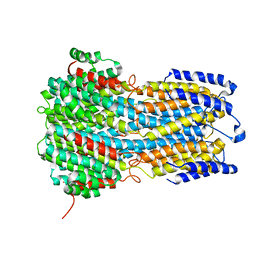 | |
1W70
 
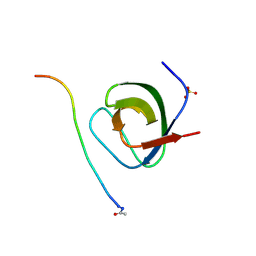 | | SH3 domain of p40phox complexed with C-terminal polyProline region of p47phox | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 1, NEUTROPHIL CYTOSOL FACTOR 4, SULFATE ION, ... | | Authors: | Massenet, C, Chenavas, S, Cohen-Addad, C, Dagher, M.-C, Brandolin, G, Pebay-Peyroula, E, Fieschi, F. | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Effects of P47Phox C-Terminus Phosphorylation on Binding Interactions with P40Phox and P67Phox: Structural and Functional Comparison of P40Phox P67Phox SH3 Domains
J.Biol.Chem., 280, 2005
|
|
5XUH
 
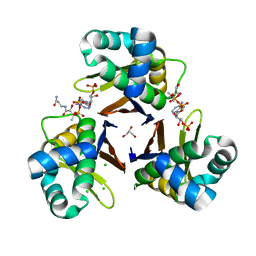 | | Crystal structure of Escherichia coli holo-[acyl-carrier-protein] synthase (AcpS) D9A mutant in complex with CoA | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Liao, Y.P, Wang, D.L, Yin, D.P, Zhang, Q.Y, Wang, Y.M, Wang, D.Q, Zhu, H.X, Chen, S. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of acyl carrier protein synthases (AcpS) from three Gram-negative bacteria
To Be Published
|
|
6IV4
 
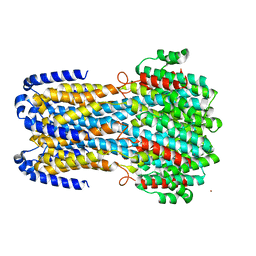 | |
7W85
 
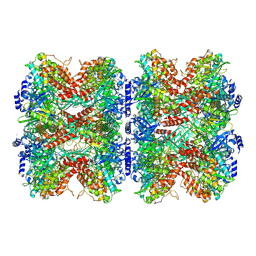 | |
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
5XUK
 
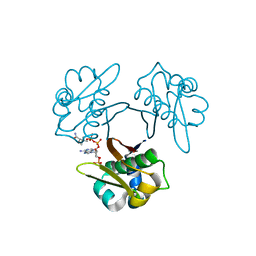 | | Crystal structure of Helicobacter pylori holo-[acyl-carrier-protein] synthase (AcpS) in complex with coenzyme A | | Descriptor: | COENZYME A, Holo-[acyl-carrier-protein] synthase | | Authors: | Liao, Y.P, Wang, D.L, Yin, D.P, Zhang, Q.Y, Wang, Y.M, Wang, D.Q, Zhu, H.X, Chen, S. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of acyl carrier protein synthases (AcpS) from three Gram-negative bacteria
To Be Published
|
|
