2Z8N
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, SULFATE ION | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1PSA
 
 | | STRUCTURE OF A PEPSIN(SLASH)RENIN INHIBITOR COMPLEX REVEALS A NOVEL CRYSTAL PACKING INDUCED BY MINOR CHEMICAL ALTERATIONS IN THE INHIBITOR | | Descriptor: | N-(ethoxycarbonyl)-L-leucyl-N-[(1R,2S,3S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methylhexyl]-L-leucinamide, PEPSIN A | | Authors: | Chen, L, Abad-Zapatero, C. | | Deposit date: | 1991-10-22 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a pepsin/renin inhibitor complex reveals a novel crystal packing induced by minor chemical alterations in the inhibitor.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
7YNK
 
 | |
7YNJ
 
 | |
7YNI
 
 | | Structure of human SGLT1-MAP17 complex bound with substrate 4D4FDG in the occluded conformation | | Descriptor: | (2R,3R,4R,5S,6R)-5-fluoranyl-6-(hydroxymethyl)oxane-2,3,4-triol, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-07-31 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of human SGLT in the occluded state reveal conformational changes during sugar transport.
Nat Commun, 14, 2023
|
|
2Z8P
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | (GLY)(GLU)(ALA)(TPO)(VAL)(PTR)(ALA), 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5TIH
 
 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | ACETATE ION, CyRPA antibody Fab Heavy Chain, CyRPA antibody Fab Light Chain, ... | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
2Z8M
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4U1Z
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U21
 
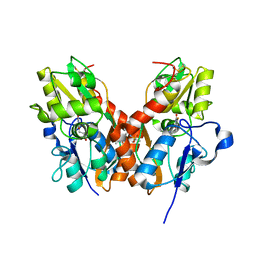 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form E | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3908 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U5G
 
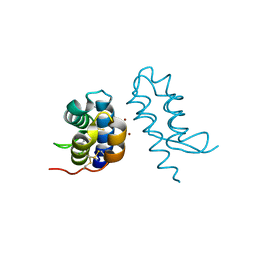 | | Crystal structure of con-ikot-ikot toxin | | Descriptor: | Con-ikot-ikot, ZINC ION | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1997 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U22
 
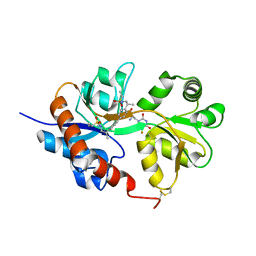 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4409 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
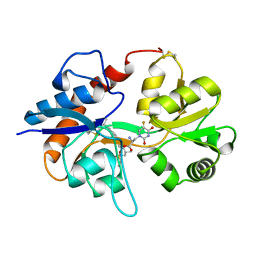 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6734 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
2DFL
 
 | | Crystal structure of left-handed RadA filament | | Descriptor: | DNA repair and recombination protein radA | | Authors: | Chen, L.T, Ko, T.P, Wang, T.F, Wang, A.H.J. | | Deposit date: | 2006-03-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins
Nucleic Acids Res., 35, 2007
|
|
4U1O
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form C | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
1WMS
 
 | | High resolution crystal structure of human Rab9 GTPase: a novel antiviral drug target | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-9A | | Authors: | Chen, L, DiGiammarino, E, Zhou, X.E, Wang, Y, Toh, D, Hodge, T.W, Meehan, E.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High resolution crystal structure of human Rab9 GTPase: A novel antiviral drug target
J.Biol.Chem., 279, 2004
|
|
7VLT
 
 | | Structure of SUR2B in complex with Mg-ATP/ADP and levcromakalim | | Descriptor: | (3S,4R)-2,2-dimethyl-3-oxidanyl-4-(2-oxidanylidenepyrrolidin-1-yl)-3,4-dihydrochromene-6-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLR
 
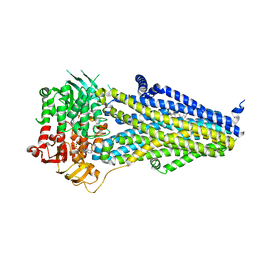 | | Structure of SUR2B in complex with Mg-ATP/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLU
 
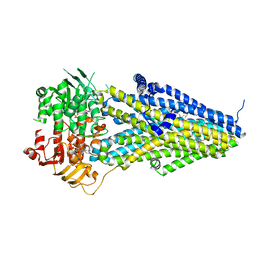 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLS
 
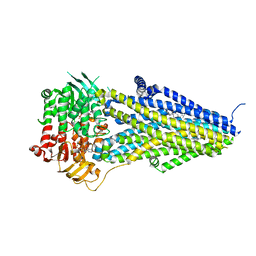 | | Structure of SUR2B in complex with MgATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
3WL0
 
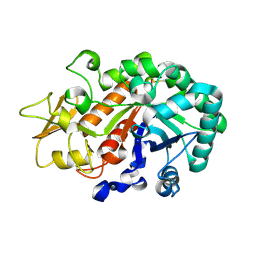 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKZ
 
 | | Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZLM
 
 | | Crystal structure of the SH3 domain of human osteoclast stimulating factor | | Descriptor: | Osteoclast stimulating factor 1 | | Authors: | Chen, L, Wang, Y, Wells, D, Toh, D, Harold, H, Zhou, J, DiGiammarino, E, Meehan, E.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure of the SH3 domain of human osteoclast-stimulating factor at atomic resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3WL1
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1DKD
 
 | |
