9CPG
 
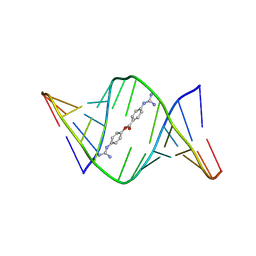 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-carbamimidamidophenyl 4-carbamimidamidobenzoate, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
9CPJ
 
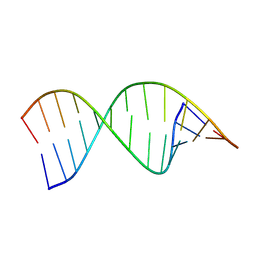 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
9CPD
 
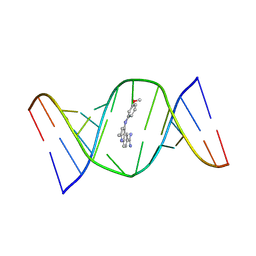 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
9CPI
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[1-(2-aminoethyl)-4,5-dihydro-1H-imidazol-2-yl]-N-[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]benzamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
9DLL
 
 | | NMR structures of small molecules bound to a model of an RNA CAG repeat expansion. | | Descriptor: | 4-carbamimidamidophenyl 4-carbamimidamidobenzoate, CAG 13-mer RNA (5'-R(*GP*AP*CP*AP*GP*CP*AP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-09-11 | | Release date: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of an RNA CAG repeat expansion.
Nucleic Acids Res., 2024
|
|
6VA3
 
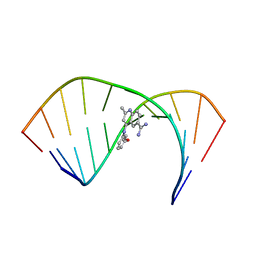 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MQC | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA2
 
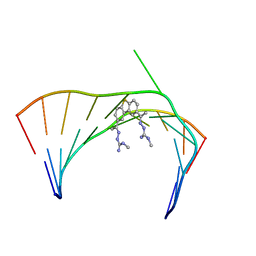 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MH5 | | Descriptor: | (2E,2'E)-2,2'-{dibenzo[b,d]thiene-2,8-diyldi[(1E)eth-1-yl-1-ylidene]}bis(N-methylhydrazine-1-carboximidamide), RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA1
 
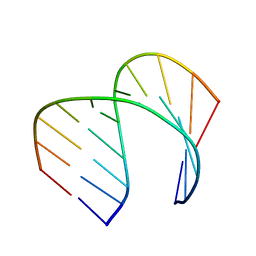 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element | | Descriptor: | RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA4
 
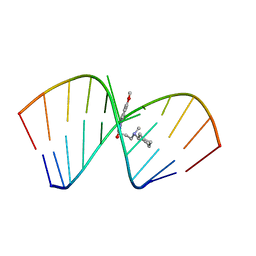 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MIP | | Descriptor: | N-[3-(8-methoxy-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-3-yl)propyl]-N-methylcyclohexanaminium, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
3KLA
 
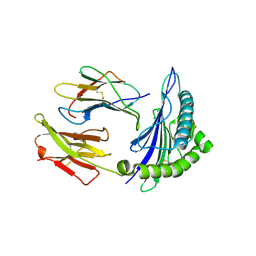 | | Ca2+ release from the endoplasmic reticulum of NY-ESO-1 specific T cells is modulated by the affinity of T cell receptor and by the use of the CD8 co-receptor | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Chen, J.L, Morgan, A.J, Stewart-Jones, G, Shepherd, D, Bossi, G, Wooldridge, L. | | Deposit date: | 2009-11-07 | | Release date: | 2010-02-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ca2+ Release from the Endoplasmic Reticulum of NY-ESO-1-Specific T Cells Is Modulated by the Affinity of TCR and by the Use of the CD8 Coreceptor.
J.Immunol., 184, 2010
|
|
5VH8
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*C)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
5VH7
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*AP*GP*CP*AP*G)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
2MXK
 
 | |
2MXL
 
 | |
2MXJ
 
 | |
