1XT3
 
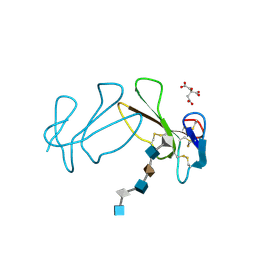 | | Structure Basis of Venom Citrate-Dependent Heparin Sulfate-Mediated Cell Surface Retention of Cobra Cardiotoxin A3 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CITRIC ACID, Cytotoxin 3 | | Authors: | Lee, S.-C, Guan, H.-H, Wang, C.-H, Huang, W.-N, Chen, C.-J, Wu, W.-G. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of citrate-dependent and heparan sulfate-mediated cell surface retention of cobra cardiotoxin A3
J.Biol.Chem., 280, 2005
|
|
4L95
 
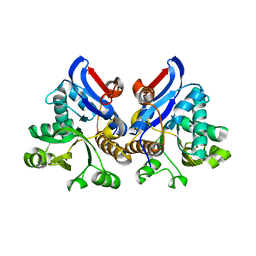 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish | | Descriptor: | GLYCEROL, Gamma-glutamyl hydrolase | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L8W
 
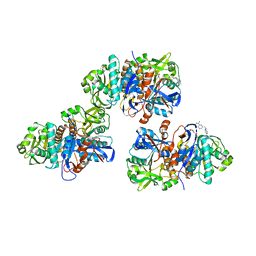 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish complex with MTX polyglutamate | | Descriptor: | D-GLUTAMIC ACID, Gamma-glutamyl hydrolase, METHOTREXATE | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L8Y
 
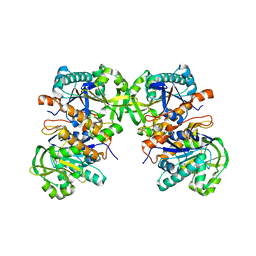 | |
4L8F
 
 | | Crystal structure of gamma-glutamyl hydrolase (C108A) complex with MTX | | Descriptor: | Gamma-glutamyl hydrolase, METHOTREXATE | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L7Q
 
 | | Crystal structure of gamma glutamyl hydrolase (wild-type) from zebrafish | | Descriptor: | GLYCEROL, Gamma-glutamyl hydrolase | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
1PKU
 
 | |
3GYX
 
 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
4HLL
 
 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 | | Descriptor: | Ankyrin(GAG)1D4 | | Authors: | Chuankhayan, P, Nangola, S, Minard, P, Boulanger, P, Hong, S.S, Tayapiwatana, C, Chen, C.-J. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Gag bioactive determinants specific to designed ankyrin and interfering in HIV-1 assembly
To be Published
|
|
6IW8
 
 | | Crystal structure of Importin-alpha and wild-type GM130 | | Descriptor: | Importin subunit alpha-1, Peptide from Golgin subfamily A member 2 | | Authors: | Chang, C.-C, Chen, C.-J, Pien, Y.-C, Tsai, S.-Y, Hsia, K.-C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Nat Commun, 10, 2019
|
|
6IWA
 
 | | Crystal structure of Importin-alpha and phosphoserine GM130 | | Descriptor: | Importin subunit alpha-1, PHOSPHOSERINE GM130 | | Authors: | Chang, C.-C, Chen, C.-J, Pien, Y.-C, Tsai, S.-Y, Hsia, K.-C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Nat Commun, 10, 2019
|
|
6K06
 
 | | Crystal structure of Importin-alpha and phosphomimetic GM130 | | Descriptor: | Importin subunit alpha-1, Peptide from Golgin subfamily A member 2 | | Authors: | Chang, C.-C, Chen, C.-J, Pien, Y.-C, Tsai, S.-Y, Hsia, K.-C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Nat Commun, 10, 2019
|
|
3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F5J
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F4V
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
7DP4
 
 | | Crystal structure of wild type Brugia malayi thymidylate synthase complexed with 2'-deoxyuridine monophosphate and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, METHOTREXATE, ... | | Authors: | Keyunratsami, K, Nualnoi, T, Wongkamchai, S, Songsiriritthigul, C, Chen, C.-J, Canyuk, B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic investigation and inhibition of Brugia malayi thymidylate synthase by a folate analog
To Be Published
|
|
7DP5
 
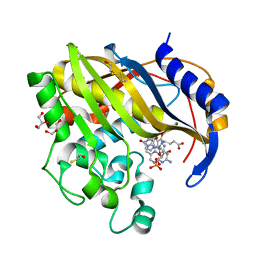 | | Crystal structure of mutant V45A Brugia malayi thymidylate synthase complexed with 2'-deoxyuridine monophosphate and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Keyunratsami, K, Nualnoi, T, Wongkamchai, S, Songsiriritthigul, C, Chen, C.-J, Canyuk, B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic investigation and inhibition of Brugia malayi thymidylate synthase by a folate analog
To Be Published
|
|
7DP6
 
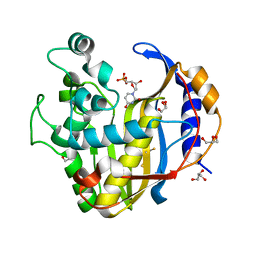 | | Crystal structure of mutant V45T Brugia malayi thymidylate synthase complexed with 2'-deoxyuridine monophosphate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Keyunratsami, K, Nualnoi, T, Wongkamchai, S, Songsiriritthigul, C, Chen, C.-J, Canyuk, B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic investigation and inhibition of Brugia malayi thymidylate synthase by a folate analog
To Be Published
|
|
7CBF
 
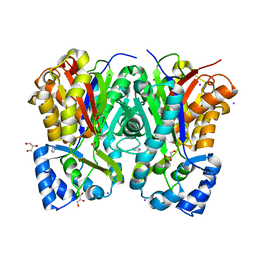 | | Crystal structure of benzophenone synthase from Garcinia mangostana L. pericarps reveals basis for substrate specificity and catalysis | | Descriptor: | 2,4,6-trihydroxybenzophenone synthase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Songsiriritthigul, C, Nualkaew, N, Chen, C.-J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of benzophenone synthase from Garcinia mangostana L. pericarps reveals basis for substrate specificity and catalysis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
