3VCX
 
 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | Descriptor: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
1U83
 
 | | PSL synthase from Bacillus subtilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphosulfolactate synthase | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PSL synthase from Bacillus subtilis
TO BE PUBLISHED
|
|
1K4N
 
 | | Structural Genomics, Protein EC4020 | | Descriptor: | Protein EC4020 | | Authors: | Zhang, R.G, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-08 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
Proteins, 51, 2003
|
|
1U0K
 
 | | The structure of a Predicted Epimerase PA4716 from Pseudomonas aeruginosa | | Descriptor: | gene product PA4716 | | Authors: | Cuff, M.E, Ginell, S.L, Rotella, F.J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hypothetical protein PA4716 from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
3O12
 
 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
3O2I
 
 | | The crystal structure of a functionally unknown protein from Leptospirillum sp. Group II UBA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of a functionally unknown protein from Leptospirillum sp. Group II UBA
TO BE PUBLISHED
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7TQ1
 
 | | Crystal structure of adaptive laboratory evolved sulfonamide-resistant Dihydropteroate Synthase (DHPS) from Escherichia coli in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Dihydropteroate synthase | | Authors: | Stogios, P.J, Skarina, T, Tan, K, Venkatesan, M, Fruci, M, Joachimiak, A, Savchenko, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
3OI7
 
 | | Structure of the structure of the H13A mutant of Ykr043C in complex with sedoheptulose-1,7-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,7-di-O-phosphono-beta-D-altro-hept-2-ulofuranose, GLYCEROL, ... | | Authors: | Singer, A.U, Xu, X, Dong, A, Cui, H, Clasquin, M.F, Caudy, A.A, Edwards, A.M, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Riboneogenesis in yeast.
Cell(Cambridge,Mass.), 145, 2011
|
|
3ON1
 
 | | The structure of a protein with unknown function from Bacillus halodurans C | | Descriptor: | BH2414 protein | | Authors: | Fan, Y, Kagan, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a protein with unknown function from Bacillus halodurans C
To be Published
|
|
1U7I
 
 | | Crystal Structure of Protein of Unknown Function PA1358 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-03 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA1358 from Pseudomonas aeruginosa
To be Published
|
|
3ON2
 
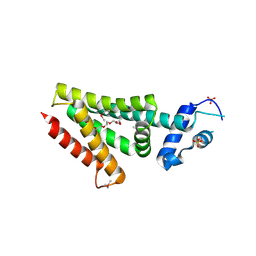 | | Structure of a protein with unknown function from Rhodococcus sp. RHA1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Probable transcriptional regulator, SULFATE ION | | Authors: | Fan, Y, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a protein with unknown function from Rhodococcus sp. RHA1
To be Published
|
|
3OKX
 
 | | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris | | Descriptor: | S-ADENOSYLMETHIONINE, YaeB-like protein RPA0152 | | Authors: | Chang, C, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris
To be Published
|
|
1U2X
 
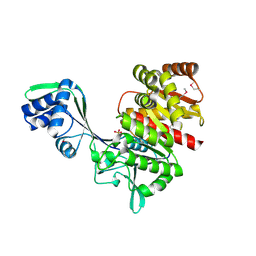 | | Crystal Structure of a Hypothetical ADP-dependent Phosphofructokinase from Pyrococcus horikoshii OT3 | | Descriptor: | ADP-specific phosphofructokinase, SULFATE ION | | Authors: | Wong, A.H.Y, Jia, Z, Skarina, T, Walker, J.R, Arrowsmith, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
1U69
 
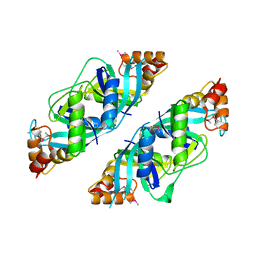 | | Crystal Structure of PA2721 Protein of Unknown Function from Pseudomonas aeruginosa PAO1 | | Descriptor: | hypothetical protein | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-21 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of a PA2721 protein from pseudomonas aeruginosa--a potential drug-resistance protein.
Proteins, 63, 2006
|
|
1KTN
 
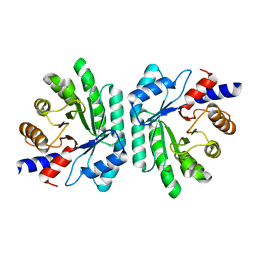 | | Structural Genomics, Protein EC1535 | | Descriptor: | 2-deoxyribose-5-phosphate aldolase | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Skarina, T, Evdokimova, E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-16 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.5A crystal structure of
2-deoxyribose-5-phosphate aldlase
To be Published
|
|
3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
6XI5
 
 | |
6XI4
 
 | |
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUM
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with paromomycin and coenzyme A | | Descriptor: | Aminocyclitol acetyltransferase ApmA, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | Descriptor: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
3MQZ
 
 | | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA. | | Descriptor: | CHLORIDE ION, GLYCEROL, uncharacterized Conserved Protein DUF1054 | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA.
To be Published
|
|
