2HMC
 
 | | The Crystal Structure of Dihydrodipicolinate Synthase DapA from Agrobacterium tumefaciens | | Descriptor: | Dihydrodipicolinate synthase, MAGNESIUM ION | | Authors: | Kim, Y, Zhang, R, Xu, X, Zheng, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Dihydrodipicolinate Synthase DapA from Agrobacterium tumefaciens
To be Published, 2006
|
|
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
3EFB
 
 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | Descriptor: | ACETIC ACID, Probable sor-operon regulator | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
2HRZ
 
 | | The crystal structure of the nucleoside-diphosphate-sugar epimerase from Agrobacterium tumefaciens | | Descriptor: | Nucleoside-diphosphate-sugar epimerase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the nucleoside-diphosphate-sugar epimerase from Agrobacterium tumefaciens
To be Published
|
|
2HWJ
 
 | | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu1540 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-01 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens
To be Published
|
|
3F6V
 
 | | Crystal structure of Possible transcriptional regulator for arsenical resistance | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator, ArsR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Possible transcriptional regulator for arsenical resistance from Rhodococcus sp.
To be Published
|
|
3I3O
 
 | | 2.06 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD-acetone | | Descriptor: | CACODYLATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 2.06 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD-acetone
To be Published
|
|
3DNH
 
 | | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58 | | Descriptor: | uncharacterized protein Atu2129 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58
To be Published
|
|
3HZN
 
 | | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase
TO BE PUBLISHED
|
|
3K67
 
 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
3OBB
 
 | | Crystal structure of a possible 3-hydroxyisobutyrate Dehydrogenase from pseudomonas aeruginosa pao1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Yakunin, A.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3E8X
 
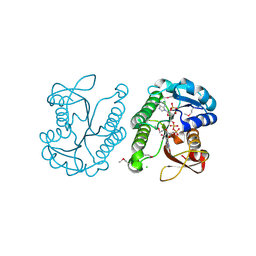 | | Putative NAD-dependent epimerase/dehydratase from Bacillus halodurans. | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NAD-dependent epimerase/dehydratase | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of putative NAD-dependent epimerase/dehydratase from Bacillus halodurans.
To be Published
|
|
3EC6
 
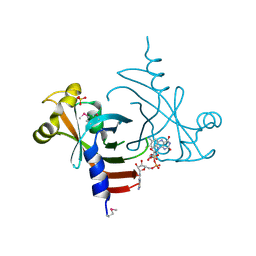 | | Crystal structure of the General Stress Protein 26 from Bacillus anthracis str. Sterne | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, General stress protein 26, SULFATE ION | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the general stress protein 26 from Bacillus anthracis str. Sterne
To be Published
|
|
3S8Y
 
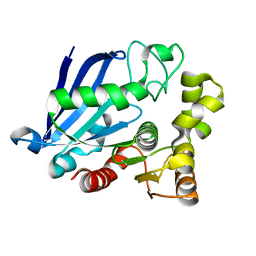 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | BROMIDE ION, Esterase APC40077 | | Authors: | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
3P48
 
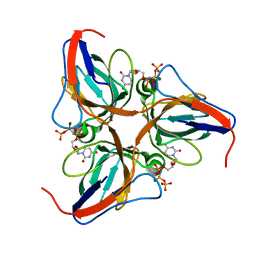 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3OJC
 
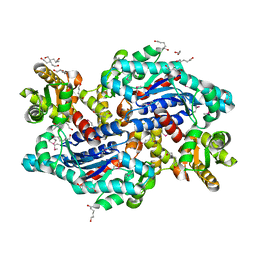 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3G25
 
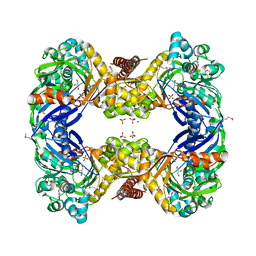 | | 1.9 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with Glycerol. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHATE ION, ... | | Authors: | Minasov, G, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with Glycerol.
TO BE PUBLISHED
|
|
3GEU
 
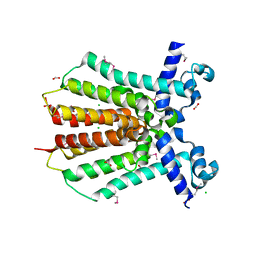 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
3QUF
 
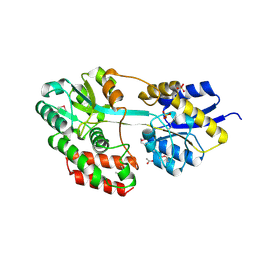 | | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | ACETIC ACID, Extracellular solute-binding protein, family 1, ... | | Authors: | Cuff, M.E, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-23 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
TO BE PUBLISHED
|
|
3S2Z
 
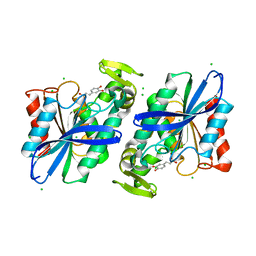 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 S106A mutant in complex with caffeic acid | | Descriptor: | CAFFEIC ACID, CHLORIDE ION, Cinnamoyl esterase | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An inserted alpha/beta subdomain shapes the catalytic pocket of Lactobacillus johnsonii cinnamoyl esterase.
Plos One, 6, 2011
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2FE7
 
 | | The crystal structure of a probable N-acetyltransferase from Pseudomonas aeruginosa | | Descriptor: | probable N-acetyltransferase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a N-acetyltransferase from Pseudomonas aeruginosa
To be Published
|
|
2FIW
 
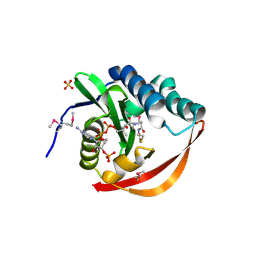 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
3BRC
 
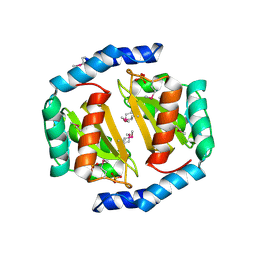 | | Crystal structure of a conserved protein of unknown function from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved protein of unknown function, PHOSPHATE ION | | Authors: | Zhang, R, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a conserved protein of unknown function from Methanobacterium thermoautotrophicum.
To be Published
|
|
2GEN
 
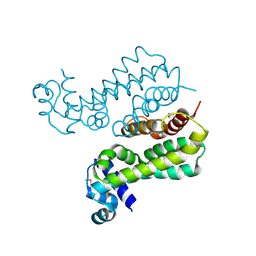 | | Structural Genomics, the crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | probable transcriptional regulator | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-20 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
