2AMI
 
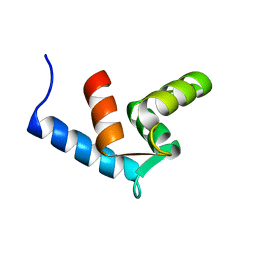 | | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | | Descriptor: | Caltractin | | Authors: | Hu, H.T, Fagan, P.A, Bunick, C.G, Sheehan, J.H, Chazin, W.J. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function.
J.Biol.Chem., 281, 2006
|
|
2BAY
 
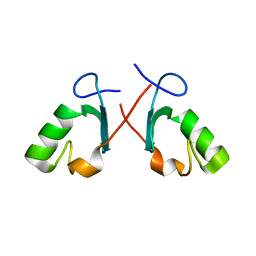 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
1CNP
 
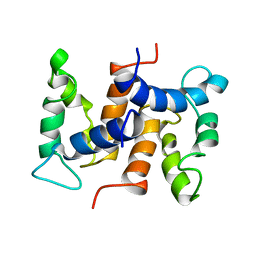 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
6DHW
 
 | |
6DU0
 
 | |
6DI6
 
 | |
6DTV
 
 | |
1PSB
 
 | | Solution structure of calcium loaded S100B complexed to a peptide from N-Terminal regulatory domain of NDR kinase. | | Descriptor: | Ndr Ser/Thr kinase-like protein, S-100 protein, beta chain | | Authors: | Bhattacharya, S, Large, E, Heizmann, C.W, Hemmings, B, Chazin, W.J. | | Deposit date: | 2003-06-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Biochemistry, 42, 2003
|
|
6DI2
 
 | |
6DTZ
 
 | | Crystal structure of eukaryotic DNA primase large subunit iron-sulfur cluster domain, Y397F mutant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, ... | | Authors: | Salay, L.E, Chazin, W.J. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Yeast require redox switching in DNA primase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1OQP
 
 | |
2L53
 
 | |
1JWD
 
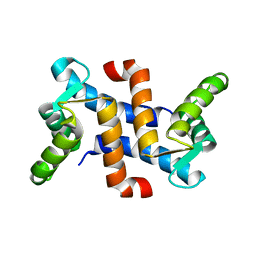 | |
2KBI
 
 | |
2KWQ
 
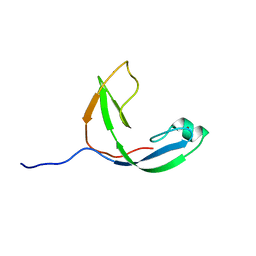 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
8UCU
 
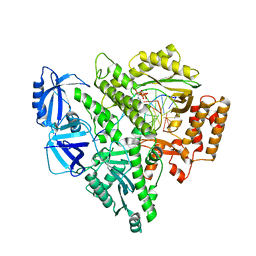 | | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U5Y
 
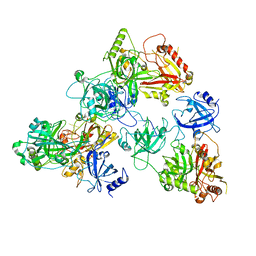 | | human RADX trimer bound to ssDNA | | Descriptor: | DNA (25-MER), RPA-related protein RADX | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Biorxiv, 2023
|
|
8V6J
 
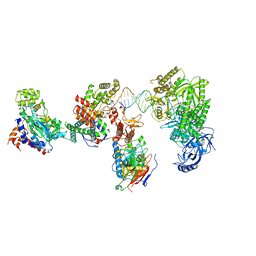 | | DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5O
 
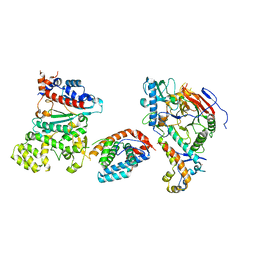 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
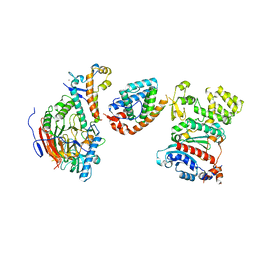 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6I
 
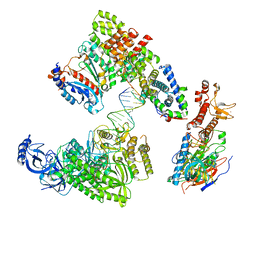 | | DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (14.06 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6G
 
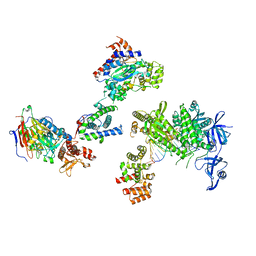 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5M
 
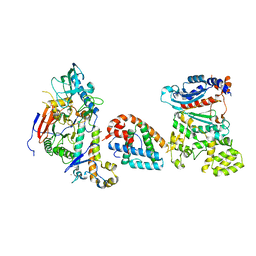 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
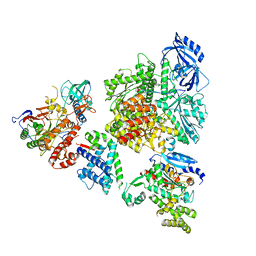 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U61
 
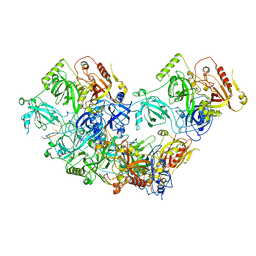 | | Human RADX tetramer bound to ssDNA | | Descriptor: | RPA-related protein RADX, dT25 DNA (25-MER) | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
