7X2Q
 
 | | Salvia miltiorrhiza CYP76AH3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sugiol synthase | | Authors: | Chang, Z. | | Deposit date: | 2022-02-26 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structure of CYP76AH3 at 3.67 Angstroms resolution
To Be Published
|
|
8WSP
 
 | | Crystal structure of SFTSV Gn and antibody SF5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab5-H, Ab5-L, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSU
 
 | | Crystal structure of SFTSV Gc and antibody | | Descriptor: | Ab-H, Ab-L, Glycoprotein C | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSN
 
 | | Crystal structure of SFTSV Gn and antibody SF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab1-H, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X1W
 
 | | CYP725A4 apo structure | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Taxadiene 5-alpha hydroxylase | | Authors: | Chang, Z, Wang, Q. | | Deposit date: | 2023-11-09 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the Catalytic Mechanism of Taxadiene-5alpha-hydroxylase from Crystallography and Computational Analyses.
Acs Catalysis, 14, 2024
|
|
8X3E
 
 | | CYP725A4-Taxa-4,11-diene complex | | Descriptor: | (1~{R},3~{R},8~{R})-4,8,12,15,15-pentamethyltricyclo[9.3.1.0^{3,8}]pentadeca-4,11-diene, PROTOPORPHYRIN IX CONTAINING FE, Taxadiene 5-alpha hydroxylase | | Authors: | Chang, Z, Wang, Q. | | Deposit date: | 2023-11-13 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unraveling the Catalytic Mechanism of Taxadiene-5alpha-hydroxylase from Crystallography and Computational Analyses.
Acs Catalysis, 14, 2024
|
|
8WVU
 
 | | Cryo-EM structure of LGR4 in complex with Rspo1 and RNF43 | | Descriptor: | E3 ubiquitin-protein ligase RNF43, Leucine-rich repeat-containing G-protein coupled receptor 4, R-spondin-1 | | Authors: | Chang, Z, Lin, C. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM structure of LGR4 in complex with Rspo1 and RNF43
To Be Published
|
|
5YM3
 
 | | CYP76AH1-4pi from salvia miltiorrhiza | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
5YLW
 
 | | CYP76AH1 from Salvia miltiorrhiza | | Descriptor: | Ferruginol synthase, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CYP76AH1 from Salvia miltiorrhiza
To Be Published
|
|
7CB9
 
 | |
7ECQ
 
 | | Crystal structure of FAM3A | | Descriptor: | Protein FAM3A, SULFATE ION, [(2R)-1-(trimethyl-$l^4-azanyl)propan-2-yl] ethanoate | | Authors: | Chang, Z, Shi, C. | | Deposit date: | 2021-03-13 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | High Resolution Crystal Structure of FAM3A shed lights on its function on beta-oxidation
To Be Published
|
|
8WVW
 
 | | Cryo-EM structure of LGR4 in state II | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4 | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of LGR4 in state I
To Be Published
|
|
8WVV
 
 | | Cryo-EM structure of LGR4 in state I | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4 | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of LGR4 in state I
To Be Published
|
|
8WVX
 
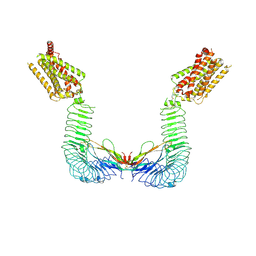 | |
8WVY
 
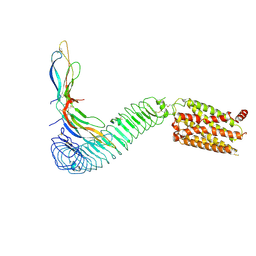 | | Cryo-EM structure of LGR4 in complex with Norrin | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4, Norrin | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of LGR4 in complex with Norrin
To Be Published
|
|
5ZUE
 
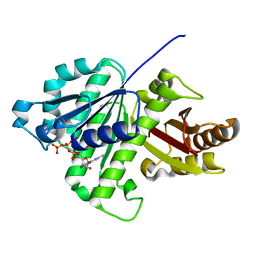 | | GTP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Guan, F, Yu, J, Yu, J, Liu, Y, Li, Y, Feng, X.H, Huang, K.C, Chang, Z, Ye, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division
Elife, 7, 2018
|
|
8Y45
 
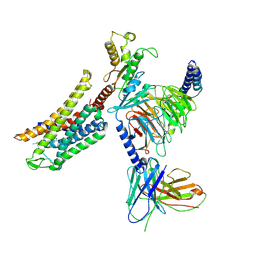 | | Cryo-EM structure of opioid receptor with biased agonist | | Descriptor: | Delta-type opioid receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2024-01-30 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of small-molecule agonist bound delta opioid receptor-G i complex enables discovery of biased compound.
Nat Commun, 15, 2024
|
|
2GAS
 
 | | Crystal Structure of Isoflavone Reductase | | Descriptor: | isoflavone reductase | | Authors: | Wang, X, He, X, Lin, J, Shao, H, Chang, Z, Dixon, R.A. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Isoflavone Reductase from Alfalfa (Medicago sativa L.)
J.Mol.Biol., 358, 2006
|
|
7FIR
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | Descriptor: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIP
 
 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIS
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
