6KYD
 
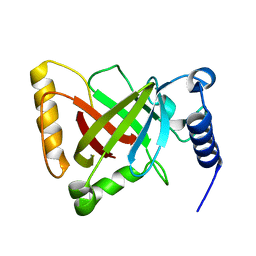 | | Structure of the R217A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
6KHE
 
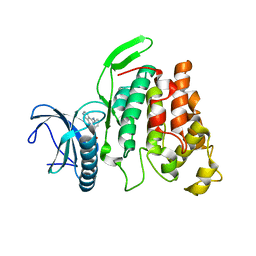 | | Crystal structure of CLK2 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KVR
 
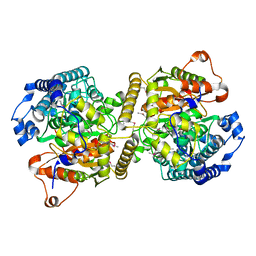 | | Fatty acid amide hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Min, C.A, Yun, J.S, Chang, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Crystals, 9, 2019
|
|
3TUD
 
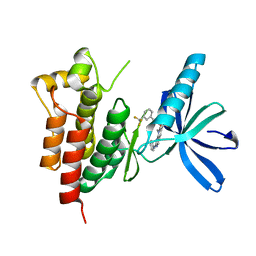 | | Crystal structure of SYK kinase domain with N-(4-methyl-3-(8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl)phenyl)-3-(trifluoromethyl)benzamide | | Descriptor: | N-{4-methyl-3-[8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]phenyl}-3-(trifluoromethyl)benzamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3UWK
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 1-methyl-6-phenyl-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, MAGNESIUM ION, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
3PQ1
 
 | | Crystal structure of human mitochondrial poly(A) polymerase (PAPD1) | | Descriptor: | Poly(A) RNA polymerase | | Authors: | Bai, Y, Srivastava, S.K, Chang, J.H, Tong, L. | | Deposit date: | 2010-11-25 | | Release date: | 2011-03-30 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase.
Mol.Cell, 41, 2011
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
3JSL
 
 | |
3AUV
 
 | | Predicting Amino Acid Preferences in the Complementarity Determining Regions of an Antibody-Antigen Recognition Interface | | Descriptor: | sc-dsFv derived from the G6-Fab | | Authors: | Yu, C.M, Peng, H.P, Chen, I.C, Lee, Y.C, Chen, J.B, Tsai, K.C, Chen, C.T, Chang, J.Y, Yang, E.W, Hsu, P.C, Jian, J.W, Hsu, H.J, Chang, H.J, Hsu, W.L, Huang, K.F, Ma, A.C, Yang, A.S. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface
Plos One, 7, 2012
|
|
3JSN
 
 | |
6JPK
 
 | |
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
6KWS
 
 | |
6KWT
 
 | |
2AOR
 
 | | Crystal structure of MutH-hemimethylated DNA complex | | Descriptor: | 5'-D(*CP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*AP*TP*CP*CP*TP*G)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
1UT9
 
 | | Structural Basis for the Exocellulase Activity of the Cellobiohydrolase CbhA from C. thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Schubot, F.D, Kataeva, I.A, Chang, J, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum.
Biochemistry, 43, 2004
|
|
4KTH
 
 | | Structure of A/Hubei/1/2010 H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Shore, D.A, Yang, H, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Antigenic Variation among Diverse Clade 2 H5N1 Viruses.
Plos One, 8, 2013
|
|
4KW1
 
 | | Structure of a/egypt/n03072/2010 h5 ha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Shore, D.A, Yang, H, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and antigenic variation among diverse clade 2 H5N1 viruses.
Plos One, 8, 2013
|
|
4KWM
 
 | | Structure of a/anhui/5/2005 h5 ha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Shore, D.A, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-24 | | Release date: | 2014-06-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and antigenic variation among diverse clade 2 H5N1 viruses.
Plos One, 8, 2013
|
|
1N6A
 
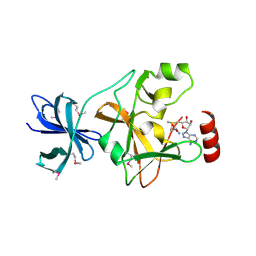 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
2L8T
 
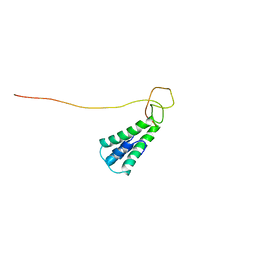 | | Staphylococcus aureus pathogenicity island 1 protein gp6, an internal scaffold in size determination | | Descriptor: | Transposon Tn557 toxic shock syndrome toxin-1 | | Authors: | Dearborn, A.D, Spilman, M.S, Damle, P.K, Chang, J.R, Monroe, E.B, Saad, J.S, Christie, G.E, Dokland, T. | | Deposit date: | 2011-01-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Staphylococcus aureus Pathogenicity Island 1 Protein gp6 Functions as an Internal Scaffold during Capsid Size Determination.
J.Mol.Biol., 412, 2011
|
|
1I91
 
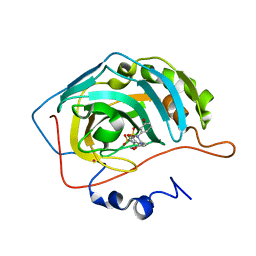 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6619 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-HYDROXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-HYDROXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I8Z
 
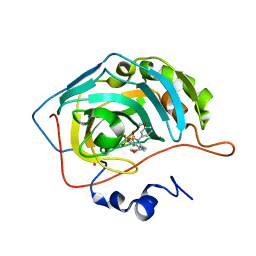 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6629 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-METHOXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-METHOXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I90
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-8520 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 4-AMINO-3,4-DIHYDRO-2-(3-METHOXYPROPYL)-, 1,1-DIOXIDE, (R) | | Descriptor: | 4-AMINO-6-[N-(3-METHOXYLPROPYL)-2H-THIENO[3,2-E][1,2]THIAZINE 1,1-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
2AOQ
 
 | | Crystal structure of MutH-unmethylated DNA complex | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*AP*TP*CP*AP*TP*GP*C)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
