2IKS
 
 | | Crystal structure of N-terminal truncated DNA-binding transcriptional dual regulator from Escherichia coli K12 | | Descriptor: | DNA-binding transcriptional dual regulator | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of N-terminal truncated DNA-binding transcriptional
dual regulator from Escherichia coli K12
To be Published
|
|
3N3T
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
2IBD
 
 | | Crystal structure of Probable transcriptional regulatory protein RHA5900 | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Probable transcriptional regulatory protein RHA5900
To be Published
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
7LZG
 
 | |
1HVR
 
 | | RATIONAL DESIGN OF POTENT, BIOAVAILABLE, NONPEPTIDE CYCLIC UREAS AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-HEXAHYDRO-5,6-DIHYDROXY-1,3-BIS[2-NAPHTHYL-METHYL]-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPIN-2-ONE | | Authors: | Chang, C.-H. | | Deposit date: | 1994-02-14 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors.
Science, 263, 1994
|
|
2IS5
 
 | |
2GE3
 
 | | Crystal structure of Probable acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | ACETYL COENZYME *A, probable acetyltransferase | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Probable acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2HWJ
 
 | | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu1540 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-01 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens
To be Published
|
|
2GRE
 
 | |
2HO5
 
 | | Crystal structure of Oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of Oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae
To be Published
|
|
2I45
 
 | |
1IHR
 
 | | Crystal structure of the dimeric C-terminal domain of TonB | | Descriptor: | BROMIDE ION, TonB protein | | Authors: | Chang, C, Mooser, A, Pluckthun, A, Wlodawer, A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold.
J.Biol.Chem., 276, 2001
|
|
1TD0
 
 | | Viral capsid protein SHP at pH 5.5 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP
J.Mol.Biol., 344, 2004
|
|
1TD3
 
 | | Crystal structure of VSHP_BPP21 in space group C2 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP.
J.Mol.Biol., 344, 2004
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2HA9
 
 | |
1TCZ
 
 | |
1N1F
 
 | | Crystal Structure of Human Interleukin-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, interleukin-19 | | Authors: | Chang, C, Magracheva, E, Kozlov, S, Fong, S, Tobin, G, Kotenko, S, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-10-17 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of interleukin-19 defines a new subfamily of helical cytokines
J.Biol.Chem., 278, 2003
|
|
2B54
 
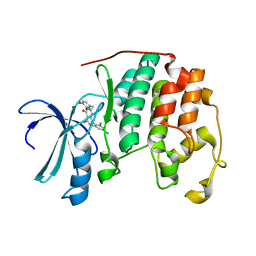 | | Human cyclin dependent kinase 2 (CKD2)complexed with DIN-232305 | | Descriptor: | 6-(3,4-DIHYDROXYBENZYL)-3-ETHYL-1-(2,4,6-TRICHLOROPHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4(5H)-ONE, Cell division protein kinase 2 | | Authors: | Chang, C.-C. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological evaluation of 1-aryl-4,5-dihydro-1h-pyraxolo[3,4-d]pyrimidin-4-one inhibitors of cyclin dependent kinases
J.Med.Chem., 47, 2004
|
|
7KA0
 
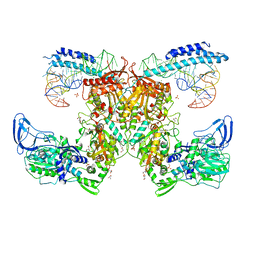 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and phenylalanine | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
7KAB
 
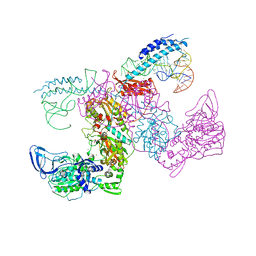 | | M. tuberculosis PheRS complex with cognate precursor tRNA and phenylalanine | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
1QBU
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R--(1ALPHA,5ALPHA,7BETA)]-3-[(CYCLOPROPHYLMETHYL)HEXAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPIN] METHYL-2-THIAZOLYLBENZAMIDE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
7QCB
 
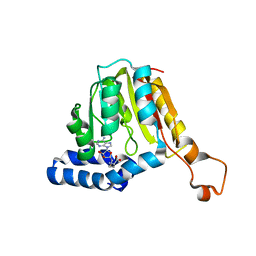 | | Williams-Beuren syndrome related methyltransferase WBSCR27 in complex with SAH | | Descriptor: | Methyltransferase-like 27, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Polshakov, V.I, Mariasina, S.S, Chang, C.-F, Efimov, S.V. | | Deposit date: | 2021-11-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Williams-Beuren Syndrome Related Methyltransferase WBSCR27: From Structure to Possible Function.
Front Mol Biosci, 9, 2022
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
