1XVS
 
 | |
1Y0E
 
 | |
3FH0
 
 | | Crystal structure of putative universal stress protein KPN_01444 - ATPase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, putative universal stress protein KPN_01444 | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative universal stress protein KPN_01444 - ATPase
To be Published
|
|
3F6V
 
 | | Crystal structure of Possible transcriptional regulator for arsenical resistance | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator, ArsR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Possible transcriptional regulator for arsenical resistance from Rhodococcus sp.
To be Published
|
|
2OPI
 
 | |
3OBF
 
 | | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302 | | Descriptor: | Putative transcriptional regulator, IclR family | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302
To be Published
|
|
1Z0X
 
 | | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583 | | Descriptor: | CHLORIDE ION, transcriptional regulator, TetR family | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583
To be Published
|
|
3OIR
 
 | | Crystal structure of sulfate transporter family protein from Wolinella succinogenes | | Descriptor: | CHLORIDE ION, FORMIC ACID, SULFATE TRANSPORTER SULFATE TRANSPORTER FAMILY PROTEIN | | Authors: | Chang, C, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of sulfate transporter family protein from Wolinella succinogenes
To be Published
|
|
1ZWY
 
 | |
3ZIQ
 
 | | minor-site specific NLS (B6) | | Descriptor: | B6NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
3ZIP
 
 | | minor-site specific NLS (A58) | | Descriptor: | A58NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
2PQV
 
 | | Crystal structure of MutT/nudix family protein from Streptococcus pneumoniae | | Descriptor: | MutT/nudix family protein, PHOSPHATE ION | | Authors: | Chang, C, Binkowski, T.A, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of MutT/nudix family protein from Streptococcus pneumoniae.
To be Published
|
|
3C0U
 
 | | Crystal structure of E.coli yaeQ protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein yaeQ | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli yaeQ protein.
To be Published
|
|
4GAK
 
 | | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale | | Descriptor: | Acyl-ACP thioesterase, CHLORIDE ION, GLYCEROL | | Authors: | Chang, C, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale
To be Published
|
|
3TEB
 
 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | Descriptor: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-12 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
3CDD
 
 | | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis | | Descriptor: | Prophage MuSo2, 43 kDa tail protein | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis.
To be Published
|
|
4G2P
 
 | | Crystal structure of peptidyl-prolyl cis-trans isomerase domain II of molecular chaperone SurA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | Chaperone SurA, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Wu, R, Adkins, J.N, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of peptidyl-prolyl cis-trans isomerase domain II of molecular chaperone SurA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S
TO BE PUBLISHED
|
|
6NBK
 
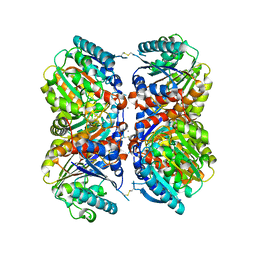 | | Crystal structure of Arginase from Bacillus cereus | | Descriptor: | Arginase, CALCIUM ION, MANGANESE (II) ION | | Authors: | Chang, C, Evdokimova, E, Mcchesney, M, Joachimiak, A, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Arginase from Bacillus cereus
To Be Published
|
|
3EFA
 
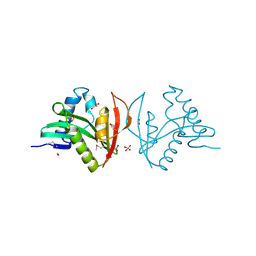 | | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chang, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum
To be Published
|
|
5E2C
 
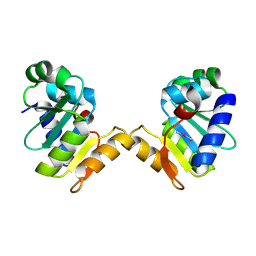 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
3T9Y
 
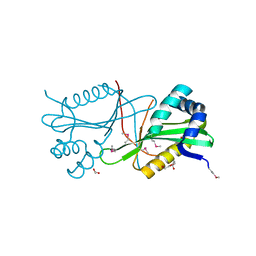 | | Crystal structure of GNAT family acetyltransferase Staphylococcus aureus subsp. aureus USA300_TCH1516 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, GNAT family, ... | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-03 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GNAT family acetyltransferase Staphylococcus aureus subsp. aureus USA300_TCH1516
To be Published
|
|
2NS0
 
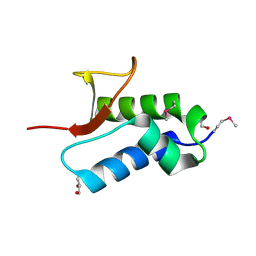 | | Crystal structure of protein RHA04536 from Rhodococcus sp | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of protein RHA04536 from Rhodococcus sp
To be Published
|
|
4EJO
 
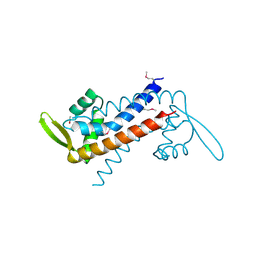 | | Crystal structure of padr family transcriptional regulator from Eggerthella lenta DSM 2243 | | Descriptor: | ETHANOL, Transcriptional regulator, PadR-like family | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of padr family transcriptional regulator from Eggerthella lenta DSM 2243
TO BE PUBLISHED
|
|
3N2Q
 
 | |
4HD1
 
 | |
