3UZO
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
4QPC
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8TWU
 
 | | Crystal structure of Cytochrome P450 AspB bound to N1-methylated cyclo-L-Trp-L-Pro | | Descriptor: | (3S,5S,8aS)-3-[(1-methyl-1H-indol-3-yl)methyl]hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450 AspB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gering, H.E, Li, X, Tang, H, Swartz, P.D, Chang, W.-C, Makris, T.M. | | Deposit date: | 2023-08-21 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Ferric-Superoxide Intermediate Initiates P450-Catalyzed Cyclic Dipeptide Dimerization.
J.Am.Chem.Soc., 145, 2023
|
|
3UYY
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
4QPD
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R8V
 
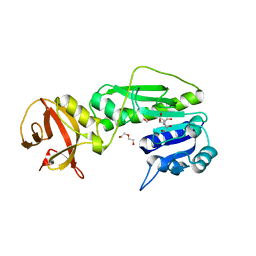 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VAY
 
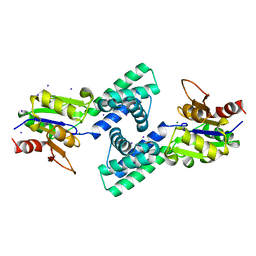 | | Crystal structure of 2-Haloacid Dehalogenase from Pseudomonas syringae pv. Tomato DC3000 | | Descriptor: | HAD-superfamily hydrolase, IODIDE ION, MAGNESIUM ION | | Authors: | Hou, Z, Zhang, H, Li, M, Chang, W. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 2-haloacid dehalogenase from Pseudomonas syringae pv. tomato DC3000
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6IS7
 
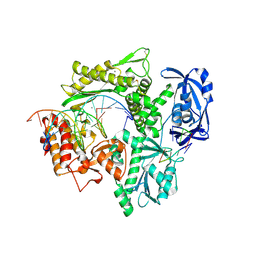 | | Structure of 9N-I DNA polymerase incorporation with dA in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*A)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISI
 
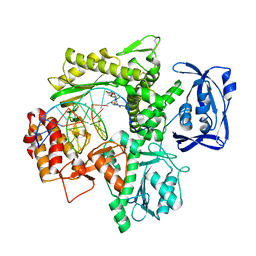 | | Structure of 9N-I DNA polymerase incorporation with 3'-CL in the active site | | Descriptor: | 3-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]propanoic acid, CALCIUM ION, DNA (5'-D(P*AP*CP*GP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISG
 
 | | Structure of 9N-I DNA polymerase incorporation with dG in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*CP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISF
 
 | | Structure of 9N-I DNA polymerase incorporation with dT in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISH
 
 | | Structure of 9N-I DNA polymerase incorporation with 3'-AL in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*(DAL))-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
1NB2
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Bacillus Halodenitrificans | | Descriptor: | Nucleoside Diphosphate Kinase | | Authors: | Chen, C.-J, Liu, M.-Y, Chang, W.-C, Chang, T, Wang, B.-C, Le Gall, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nucleoside diphosphate kinase from Bacillus halodenitrificans: coexpression of its activity with a Mn-superoxide dismutase.
J.Struct.Biol., 142, 2003
|
|
8XID
 
 | |
5GKB
 
 | | Crystal Structure of Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster without citrate inside | | Descriptor: | Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
5GGE
 
 | | Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster | | Descriptor: | CITRIC ACID, Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
1RWT
 
 | | Crystal Structure of Spinach Major Light-harvesting complex at 2.72 Angstrom Resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Liu, Z, Yan, H, Wang, K, Kuang, T, Zhang, J, Gui, L, An, X, Chang, W. | | Deposit date: | 2003-12-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of spinach major light-harvesting complex at 2.72 A resolution
Nature, 428, 2004
|
|
6L6W
 
 | | The structure of ScoE with intermediate | | Descriptor: | (3R)-3-[[(1R)-1,2-bis(oxidanyl)-2-oxidanylidene-ethyl]amino]butanoic acid, FE (II) ION, FORMIC ACID, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1LIA
 
 | |
1TGR
 
 | | Crystal Structure of mini-IGF-1(2) | | Descriptor: | Insulin-like growth factor IA | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2004-05-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42A crystal structure of mini-IGF-1(2): an analysis of the disulfide isomerization property and receptor binding property of IGF-1 based on the three-dimensional structure
Biochem.Biophys.Res.Commun., 326, 2004
|
|
5HTP
 
 | |
5HU2
 
 | |
5HTY
 
 | |
