1I9L
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(4-FLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(4-FLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1S1G
 
 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1I9O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I9Q
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(3,4,5-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(3,4,5-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1EZT
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-11 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
4G91
 
 | | CCAAT-binding complex from Aspergillus nidulans | | Descriptor: | HAPB protein, HapE, Transcription factor HapC (Eurofung) | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
1NON
 
 | | PyrR, the regulator of the pyrimidine biosynthetic operon in Bacillus caldolyticus | | Descriptor: | PyrR bifunctional protein | | Authors: | Switzer, R.L, Chander, P, Smith, J.L, Halbig, K.M, Miller, J.K, Bonner, H.K, Grabner, G.K. | | Deposit date: | 2003-01-16 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides
J.Bacteriol., 187, 2005
|
|
1FM1
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
1FLS
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
4G92
 
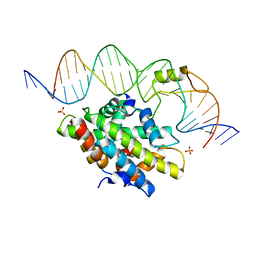 | | CCAAT-binding complex from Aspergillus nidulans with DNA | | Descriptor: | DNA, HAPB protein, HapE, ... | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
1EZY
 
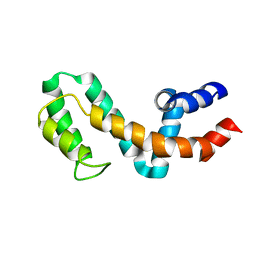 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-12 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
2K0Q
 
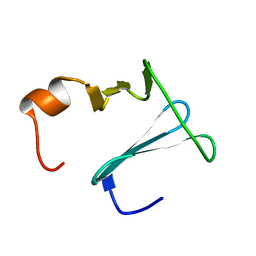 | | Solution structure of CopK, a periplasmic protein involved in copper resistance in Cupriavidus metallidurans CH34 | | Descriptor: | Putative uncharacterized protein copK | | Authors: | Bersch, B, Favier, A, Schanda, P, Coves, J, van Aelst, S, Vallaeys, T, Wattiez, R, Mergeay, M. | | Deposit date: | 2008-02-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular structure and metal-binding properties of the periplasmic CopK protein expressed in Cupriavidus metallidurans CH34 during copper challenge.
J.Mol.Biol., 380, 2008
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
6HWN
 
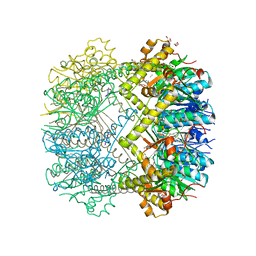 | | Structure of Thermus thermophilus ClpP in complex with a tripeptide. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, Unknown tripeptide | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
6HWM
 
 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
2LXS
 
 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2LXT
 
 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Cyclic AMP-responsive element-binding protein 1, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2L3Z
 
 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
