1AOR
 
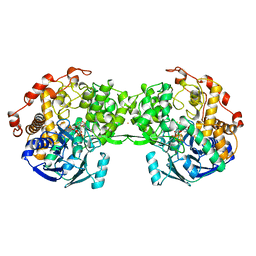 | | STRUCTURE OF A HYPERTHERMOPHILIC TUNGSTOPTERIN ENZYME, ALDEHYDE FERREDOXIN OXIDOREDUCTASE | | Descriptor: | ALDEHYDE FERREDOXIN OXIDOREDUCTASE, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Chan, M.K, Mukund, S, Kletzin, A, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1995-02-13 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase.
Science, 267, 1995
|
|
1DFF
 
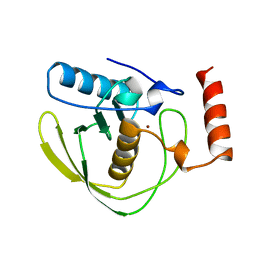 | | PEPTIDE DEFORMYLASE | | Descriptor: | PEPTIDE DEFORMYLASE, ZINC ION | | Authors: | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | Deposit date: | 1997-08-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
4XQK
 
 | |
5ELX
 
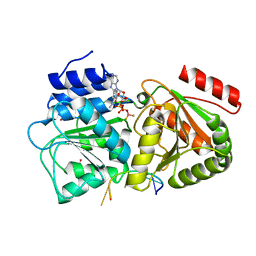 | | S. cerevisiae Dbp5 bound to RNA and mant-ADP BeF3 | | Descriptor: | ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Merchant, M.K, Modis, Y. | | Deposit date: | 2015-11-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Pi Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5).
J.Mol.Biol., 428, 2016
|
|
4E4G
 
 | | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
4E6M
 
 | | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium)
To be Published
|
|
1L2Q
 
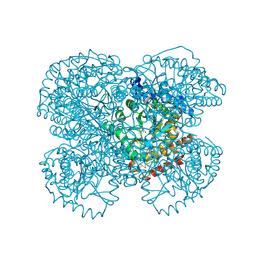 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
1LSW
 
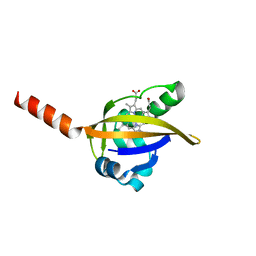 | | Crystal structure of the ferrous BjFixL heme domain | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-20 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
1LT0
 
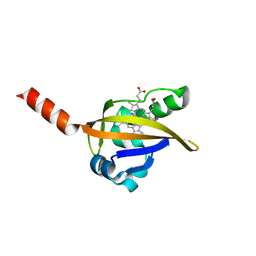 | | Crystal structure of the CN-bound BjFixL heme domain | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-20 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
1LSV
 
 | | Crystal structure of the CO-bound BjFixL heme domain | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-19 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
1LSX
 
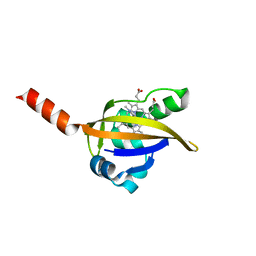 | | Crystal structure of the methylimidazole-bound BjFixL heme domain | | Descriptor: | 1-METHYLIMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-20 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
3HQ2
 
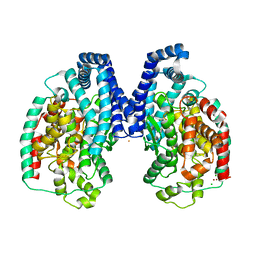 | | BsuCP Crystal Structure | | Descriptor: | Bacillus subtilis M32 carboxypeptidase, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Lee, M.M, Isaza, C.E, White, J.D, Chen, R.P.-Y, Liang, G.F.-C, He, H.T.-F, Chan, S.I, Chan, M.K. | | Deposit date: | 2009-06-05 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the substrate length restriction of M32 carboxypeptidases: Characterization of two distinct subfamilies.
Proteins, 77, 2009
|
|
1BSJ
 
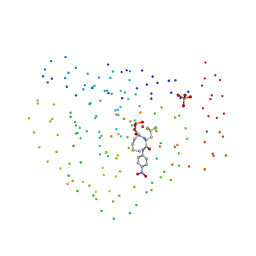 | | COBALT DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
3HOA
 
 | |
4RPF
 
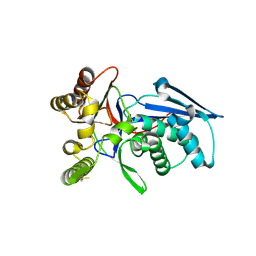 | | Crystal structure of homoserine kinase from Yersinia pestis Nepal516, NYSGRC target 032715 | | Descriptor: | CITRIC ACID, Homoserine kinase | | Authors: | Ptskovsky, Y, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Homoserine Kinase from Yersinia Pestis Nepal516, Nysgrc Target 032715
To be Published
|
|
4TYM
 
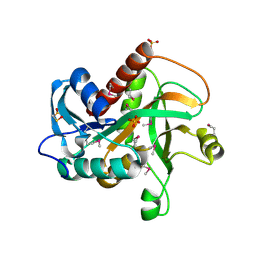 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
4WR2
 
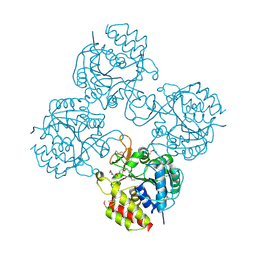 | | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Pyrimidine-specific ribonucleoside hydrolase RihA | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site
To be published
|
|
5DK6
 
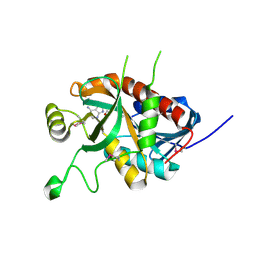 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
8CYH
 
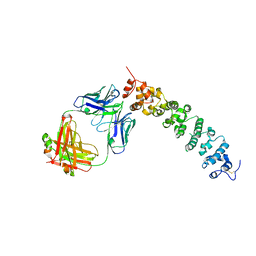 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
7XCL
 
 | |
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
