2PWQ
 
 | | Crystal structure of a putative ubiquitin conjugating enzyme from Plasmodium yoelii | | Descriptor: | Ubiquitin conjugating enzyme | | Authors: | Qiu, W, Dong, A, Hassanali, A, Lin, L, Brokx, S, Altamentova, S, Hills, T, Lew, J, Ravichandran, M, Kozieradzki, I, Zhao, Y, Schapira, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative ubiquitin conjugating enzyme from Plasmodium yoelii.
To be Published
|
|
7C6R
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellopentaose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C69
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to sophorose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Sugar ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
3DM1
 
 | | Crystal structure of the complex of human chromobox homolog 3 (CBX3) with peptide | | Descriptor: | Chromobox protein homolog 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
3PNW
 
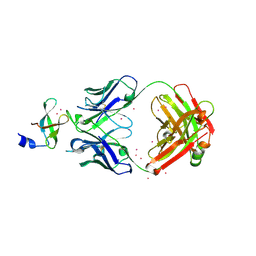 | | Crystal Structure of the tudor domain of human TDRD3 in complex with an anti-TDRD3 FAB | | Descriptor: | FAB heavy chain, FAB light chain, Tudor domain-containing protein 3, ... | | Authors: | Loppnau, P, Tempel, W, Wernimont, A.K, Lam, R, Ravichandran, M, Adams-Cioaba, M.A, Persson, H, Sidhu, S.S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CDR-H3 Diversity Is Not Required for Antigen Recognition by Synthetic Antibodies.
J.Mol.Biol., 425, 2013
|
|
3DZO
 
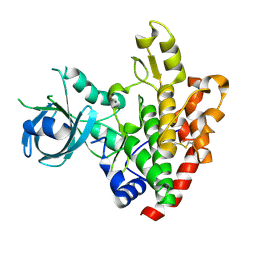 | | Crystal structure of a rhoptry kinase from toxoplasma gondii | | Descriptor: | MAGNESIUM ION, Rhoptry kinase domain | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Ni, S, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sibley, D, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
1EJJ
 
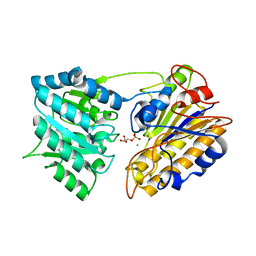 | | CRYSTAL STRUCTURAL ANALYSIS OF PHOSPHOGLYCERATE MUTASE COCRYSTALLIZED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus.
EMBO J., 19, 2000
|
|
1TUE
 
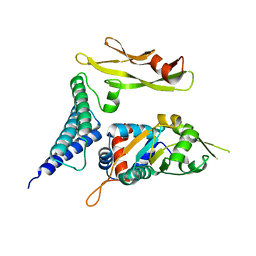 | |
2XM3
 
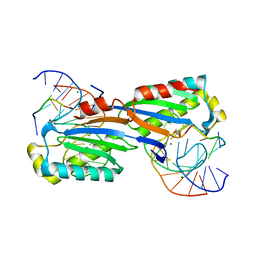 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | Descriptor: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
3PHD
 
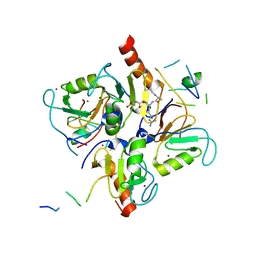 | | Crystal structure of human HDAC6 in complex with ubiquitin | | Descriptor: | Histone deacetylase 6, Polyubiquitin, ZINC ION | | Authors: | Dong, A, Qui, W, Ravichandran, M, Schuetz, A, Loppnau, P, Li, F, Mackenzie, F, Kozieradzki, I, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Aggregates Are Recruited to Aggresome by Histone Deacetylase 6 via Unanchored Ubiquitin C Termini.
J.Biol.Chem., 287, 2012
|
|
6BJ9
 
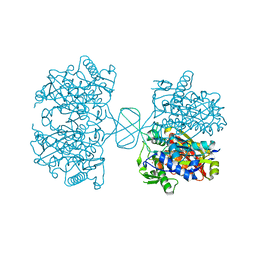 | |
6BJA
 
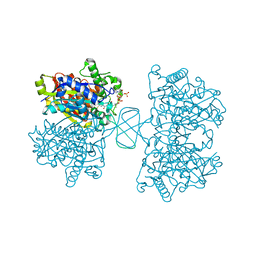 | |
3C5K
 
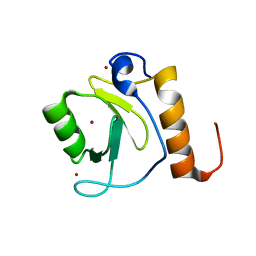 | | Crystal structure of human HDAC6 zinc finger domain | | Descriptor: | Histone deacetylase 6, ZINC ION | | Authors: | Dong, A, Ravichandran, M, Schuetz, A, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human HDAC6 zinc finger domain.
To be Published
|
|
6BJB
 
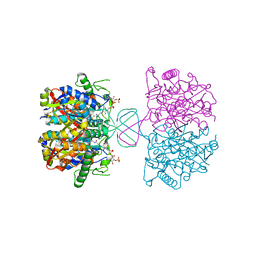 | |
7C14
 
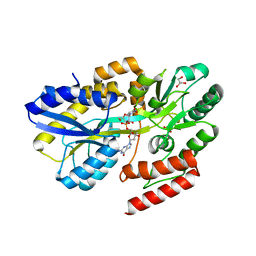 | | Crystal structure of a dinucleotide-binding protein (Q274A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form I) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0Z
 
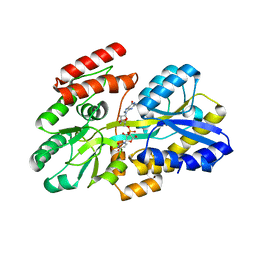 | | Crystal structure of a dinucleotide-binding protein (Y246A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, Sugar ABC transporter, periplasmic sugar-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C1B
 
 | | Crystal structure of a dinucleotide-binding protein (F79A/Y224A/Y246A and deletion of residues 50-75) of ABC transporter (unbound form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sugar ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0W
 
 | | Crystal structure of a dinucleotide-binding protein (Y224A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, GLYCEROL, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
3DU7
 
 | | Tubulin-colchicine-phomopsin A: Stathmin-like domain complex | | Descriptor: | 2-MERCAPTO-N-[1,2,3,10-TETRAMETHOXY-9-OXO-5,6,7,9-TETRAHYDRO-BENZO[A]HEPTALEN-7-YL]ACETAMIDE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cormier, A, Marchand, M, Ravelli, R.B, Knossow, M, Gigant, B. | | Deposit date: | 2008-07-17 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural insight into the inhibition of tubulin by vinca domain peptide ligands
Embo Rep., 9, 2008
|
|
1EQJ
 
 | | CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J.Biol.Chem., 275, 2000
|
|
5C1S
 
 | |
5C1T
 
 | | Crystal structure of the GTP-bound wild type EhRabX3 from Entamoeba histolytica | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Small GTPase EhRabX3 | | Authors: | Srivastava, V.K, Chandra, M, Datta, S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal Structure Analysis of Wild Type and Fast Hydrolyzing Mutant of EhRabX3, a Tandem Ras Superfamily GTPase from Entamoeba histolytica.
J.Mol.Biol., 428, 2016
|
|
1SPY
 
 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-FREE STATE, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN C | | Authors: | Spyracopoulos, L, Li, M.X, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
3TZD
 
 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | Descriptor: | Chromobox protein homolog 3, Histone H1.4 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-07 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
2XMA
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
