8TR3
 
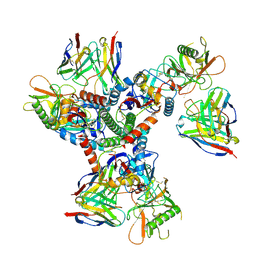 | | Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CNE40 SOSIP Envelope glycoprotein gp120, ... | | Authors: | Chan, K.-W, Kong, X.P. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human CD4-binding site antibody elicited by polyvalent DNA prime-protein boost vaccine neutralizes cross-clade tier-2-HIV strains.
Nat Commun, 15, 2024
|
|
7SGN
 
 | |
7SGO
 
 | |
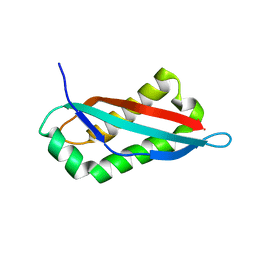
7SGP
 
 | |
6YSS
 
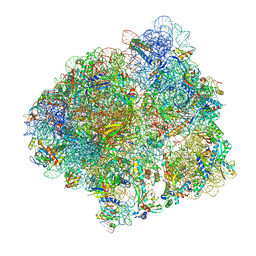 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YST
 
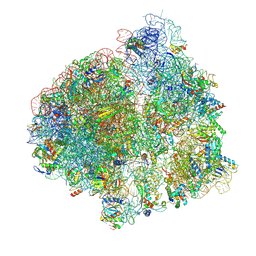 | | Structure of the P+9 ArfB-ribosome complex with P/E hybrid tRNA in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSU
 
 | | Structure of the P+0 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
7RAI
 
 | |
6VY2
 
 | |
5O38
 
 | | Human Brd2(BD2) mutant in free form | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, 3-[(2~{R})-2-oxidanylpropoxy]-2-[[(2~{R})-2-oxidanylpropoxy]methyl]-2-[[(2~{S})-2-oxidanylpropoxy]methyl]propan-1-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3I
 
 | | Human Brd2(BD2) mutant in complex with AL-tBu | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3D
 
 | | Human Brd2(BD2) mutant in complex with 9-ET | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3A
 
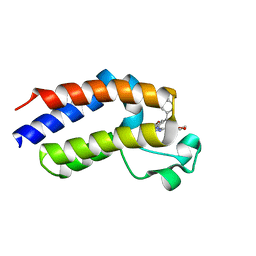 | | Human Brd2(BD2) mutant in complex with ET | | Descriptor: | Bromodomain-containing protein 2, methyl (2R)-2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]butanoate | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3E
 
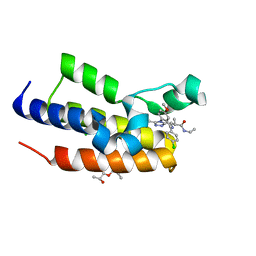 | | Human Brd2(BD2) mutant in complex with Me-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-propanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3G
 
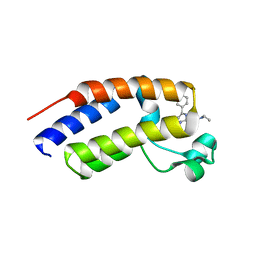 | | Human Brd2(BD2) mutant in complex with AL-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-pent-4-enamide, Bromodomain-containing protein 2 | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3H
 
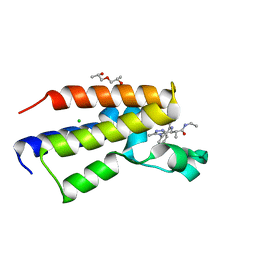 | | Human Brd2(BD2) mutant in complex with 9-ME-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-9-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-propanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
6DB5
 
 | |
6DB7
 
 | |
6DB6
 
 | |
3PQC
 
 | | Crystal structure of Thermotoga maritima ribosome biogenesis GTP-binding protein EngB (YsxC/YihA) in complex with GDP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, Probable GTP-binding protein engB | | Authors: | Chan, K.H, Wong, K.B. | | Deposit date: | 2010-11-26 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of an essential GTPase, YsxC, from Thermotoga maritima
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PR1
 
 | |
2VW9
 
 | | Single stranded DNA binding protein complex from Helicobacter pylori | | Descriptor: | POLY-DT, SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Chan, K.-W, Wang, C.-H, Lee, Y.-J, Sun, Y.-J. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single-Stranded DNA-Binding Protein Complex from Helicobacter Pylori Suggests an Ssdna-Binding Surface.
J.Mol.Biol., 388, 2009
|
|
8WT1
 
 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
7Q5N
 
 | | Crystal structure of Chaetomium thermophilum Ahp1-Urm1 complex | | Descriptor: | Thioredoxin domain-containing protein, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | Deposit date: | 2021-11-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
