2F8N
 
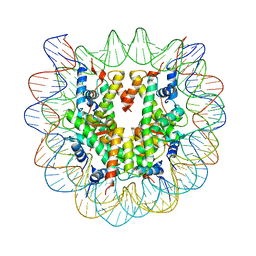 | |
1U35
 
 | | Crystal structure of the nucleosome core particle containing the histone domain of macroH2A | | Descriptor: | H2A histone family, Hist1h4i protein, Histone H3.1, ... | | Authors: | Chakravarthy, S, Gundimella, S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of the histone variant macroH2A.
Mol.Cell.Biol., 25, 2005
|
|
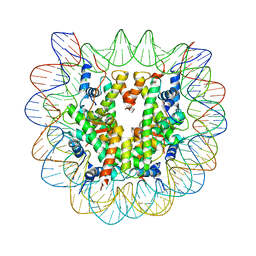
1YD9
 
 | | 1.6A Crystal Structure of the Non-Histone Domain of the Histone Variant MacroH2A1.1. | | Descriptor: | Core histone macro-H2A.1, GOLD ION | | Authors: | Chakravarthy, S, Swamy, G.Y.S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the histone variant macroH2A
Mol.Cell.Biol., 25, 2005
|
|
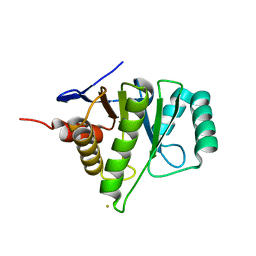
4MN3
 
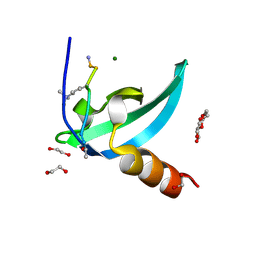 | | Chromodomain antagonists that target the polycomb-group methyllysine reader protein Chromobox homolog 7 (CBX7) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Chromobox protein homolog 7, ... | | Authors: | Chakravarthi, S, Daze, K, Douglas, S, Quon, T, Dev, A, Peng, F, Heller, M, Boulanger, M.J, Wulff, J, Hof, F. | | Deposit date: | 2013-09-09 | | Release date: | 2014-04-02 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | Chromodomain Antagonists That Target the Polycomb-Group Methyllysine Reader Protein Chromobox Homolog 7 (CBX7).
J.Med.Chem., 57, 2014
|
|
2NQB
 
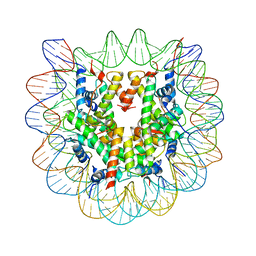 | | Drosophila Nucleosome Structure | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Luger, K, Chakravarthy, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative analysis of nucleosome structures from different species.
To be Published
|
|
6NXJ
 
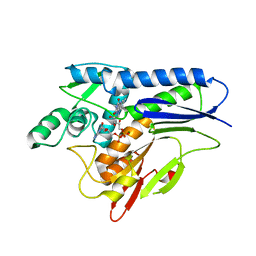 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NXI
 
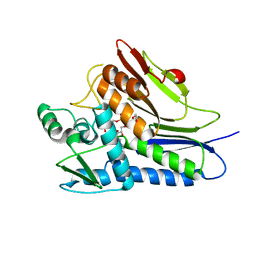 | | Flavin Transferase ApbE from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
5VHG
 
 | | Crystal structure of pentad mutant GAPR-1 | | Descriptor: | Golgi-associated plant pathogenesis-related protein 1, SULFATE ION | | Authors: | Li, Y, Zhao, Y, Su, M, Chakravarthy, S, Colbert, C.L, Levine, B, Sinha, S.C. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural insights into the interaction of the conserved mammalian proteins GAPR-1 and Beclin 1, a key autophagy protein.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4C0J
 
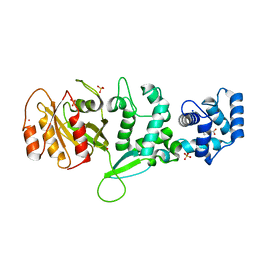 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains in the apo state (Apo-MiroS) | | Descriptor: | L-HOMOSERINE, MITOCHONDRIAL RHO GTPASE, SODIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
4C0K
 
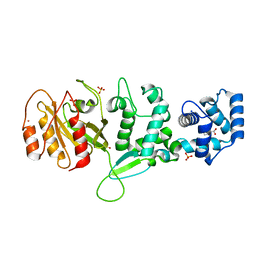 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains bound to one calcium ion (Ca-MiroS) | | Descriptor: | CALCIUM ION, L-HOMOSERINE, MITOCHONDRIAL RHO GTPASE, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
4C0L
 
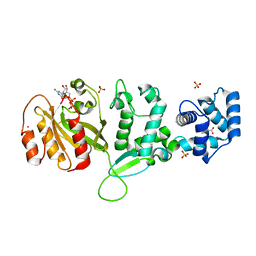 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains bound to one magnesium ion and Mg:GDP (MgGDP-MiroS) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, L-HOMOSERINE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
5KN9
 
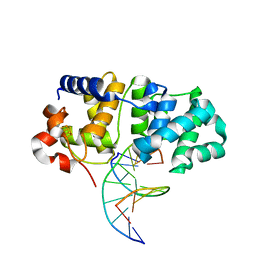 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
5KN8
 
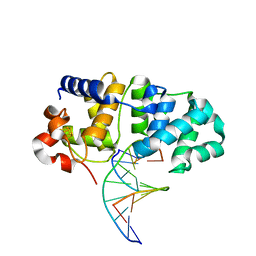 | | MutY N-terminal domain in complex with undamaged DNA | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
