6MBI
 
 | |
7MZV
 
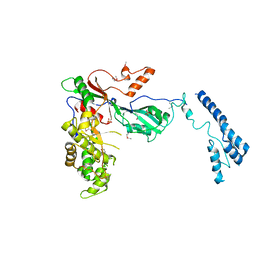 | | Structure of yeast pseudouridine synthase 7 (PUS7) | | 分子名称: | Multisubstrate pseudouridine synthase 7, SULFATE ION | | 著者 | Purchal, M, Koutmos, M. | | 登録日 | 2021-05-24 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5DJW
 
 | |
5F7C
 
 | |
5OVY
 
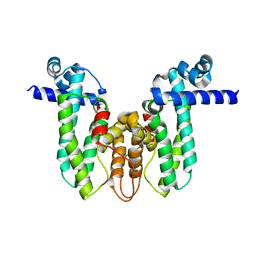 | | Crystal structure of MAB_4384 tetR | | 分子名称: | Putative transcriptional regulator, TetR family | | 著者 | Richard, M, Gutierrez, A.V, Viljoen, A, Ghigo, E, Blaise, M, Kremer, L. | | 登録日 | 2017-08-30 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanistic and Structural Insights Into the Unique TetR-Dependent Regulation of a Drug Efflux Pump inMycobacterium abscessus.
Front Microbiol, 9, 2018
|
|
4Z66
 
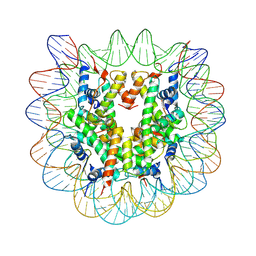 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | 登録日 | 2015-04-03 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
6HHC
 
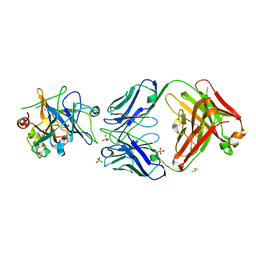 | | Allosteric Inhibition as a new mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated form of Coagulation Factor XI | | 分子名称: | Coagulation factor XI, DIMETHYL SULFOXIDE, FXIA ANTIBODY FAB HEAVY CHAIN, ... | | 著者 | Schaefer, M, Buchmueller, A, Dittmer, F, Strassburger, J, Wilmen, A. | | 登録日 | 2018-08-27 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Allosteric Inhibition as a New Mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated Form of Coagulation Factor XI.
J.Mol.Biol., 431, 2019
|
|
5G5R
 
 | |
5ELX
 
 | |
5G5X
 
 | |
7FBY
 
 | | Crystal Structure of PH0140 from Pyrococcus horikosii OT3 | | 分子名称: | 1,2-ETHANEDIOL, ISOLEUCINE, Transcriptional regulatory protein | | 著者 | Richard, M, Ahmad, M, Pal, R.K, Biswal, B.K, Jeyakanthan, J. | | 登録日 | 2021-07-13 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal Structure of PH0140 from Pyrococcus horikosii OT3
To Be Published
|
|
5IG5
 
 | |
5IG4
 
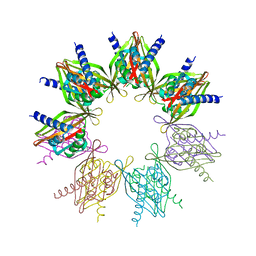 | | Crystal structure of N. vectensis CaMKII-A hub | | 分子名称: | GLYCEROL, Predicted protein | | 著者 | Bhattacharyya, M, Pappireddi, N, Gee, C.L, Barros, T, Kuriyan, J. | | 登録日 | 2016-02-26 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG1
 
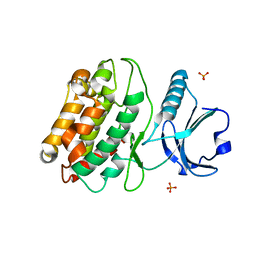 | | Crystal structure of S. rosetta CaMKII kinase domain | | 分子名称: | CAMK/CAMK2 protein kinase, PHOSPHATE ION | | 著者 | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | 登録日 | 2016-02-26 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
7Z90
 
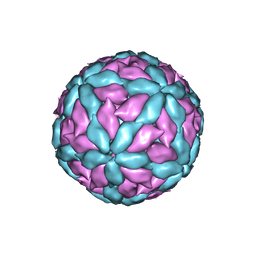 | |
5IG0
 
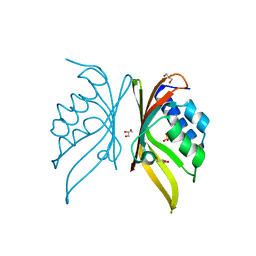 | | Crystal structure of S. rosetta CaMKII hub | | 分子名称: | CAMK/CAMK2 protein kinase, GLYCEROL, SULFATE ION | | 著者 | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | 登録日 | 2016-02-26 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5G2N
 
 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | 分子名称: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | 著者 | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | 登録日 | 2016-04-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
5G1X
 
 | | Crystal structure of Aurora-A kinase in complex with N-Myc | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, MAGNESIUM ION, ... | | 著者 | Richards, M.W, Burgess, S.G, Bayliss, R. | | 登録日 | 2016-03-31 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural Basis of N-Myc Binding by Aurora-A and its Destabilization by Kinase Inhibitors
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ID5
 
 | |
5A0R
 
 | | Product peptide-bound structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | 分子名称: | GLYCEROL, PRODUCT PEPTIDE, ZINC ION, ... | | 著者 | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | 登録日 | 2015-04-22 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.251 Å) | | 主引用文献 | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | 分子名称: | Suppressor of Fused | | 著者 | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | 登録日 | 2002-06-19 | | 公開日 | 2004-02-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
5A0P
 
 | | Apo-structure of metalloprotease Zmp1 from Clostridium difficile | | 分子名称: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | 著者 | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | 登録日 | 2015-04-22 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0X
 
 | | Substrate peptide-bound structure of metalloprotease Zmp1 variant E143AY178F from Clostridium difficile | | 分子名称: | SUBSTRATE PEPTIDE, ZINC ION, ZINC METALLOPROTEASE ZMP1 | | 著者 | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | 登録日 | 2015-04-23 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
3MG0
 
 | | Structure of yeast 20S proteasome with bortezomib | | 分子名称: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome component C1, Proteasome component C11, ... | | 著者 | Sintchak, M.D. | | 登録日 | 2010-04-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3MG4
 
 | | Structure of yeast 20S proteasome with Compound 1 | | 分子名称: | (2S)-2-amino-N-[(1S)-1-({(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}carbamoyl)-3-phenylpropyl]-4-phenylbutanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | 著者 | Sintchak, M.D. | | 登録日 | 2010-04-05 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
