6KVR
 
 | | Fatty acid amide hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Min, C.A, Yun, J.S, Chang, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Crystals, 9, 2019
|
|
5XR4
 
 | | Crystal structure of RabA1a in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a, ... | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
6KWS
 
 | |
5XR7
 
 | | Crystal structure of RabA1a (Q72K) in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
6KWT
 
 | |
5XR6
 
 | | Crystal structure of RabA1a in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
7ELF
 
 | |
7E6H
 
 | | glucose-6-phosphate dehydrogenase from Kluyveromyces lactis | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Ha, V.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
7E6I
 
 | | Glucose-6-phosphate dehydrogenase in complex with its substrate glucose-6-phosphate | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Vu, H.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
6TGB
 
 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6TGC
 
 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
5BY2
 
 | |
4J7N
 
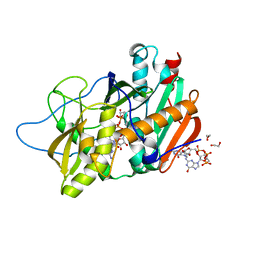 | | Crystal structure of mouse DXO in complex with M7GPPPG cap | | Descriptor: | 1,2-ETHANEDIOL, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 9-METHYLGUANINE, ... | | Authors: | Kilic, T, Chang, J.H, Tong, L. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing.
Mol.Cell, 50, 2013
|
|
3PQ1
 
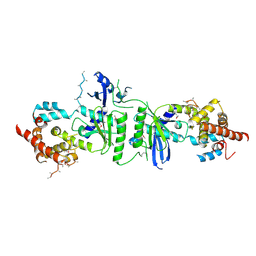 | | Crystal structure of human mitochondrial poly(A) polymerase (PAPD1) | | Descriptor: | Poly(A) RNA polymerase | | Authors: | Bai, Y, Srivastava, S.K, Chang, J.H, Tong, L. | | Deposit date: | 2010-11-25 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase.
Mol.Cell, 41, 2011
|
|
6IQ1
 
 | |
2ZU6
 
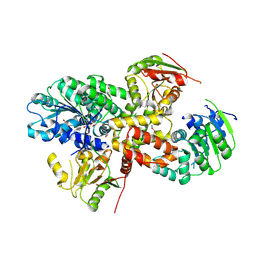 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
1N4M
 
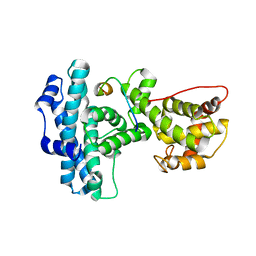 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
1N6A
 
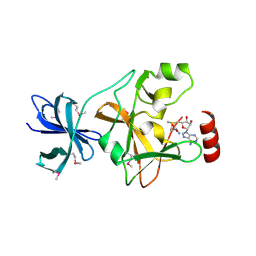 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6C
 
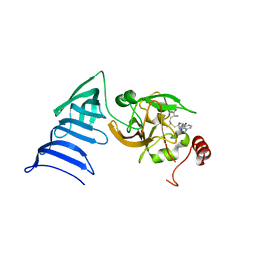 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
5ELL
 
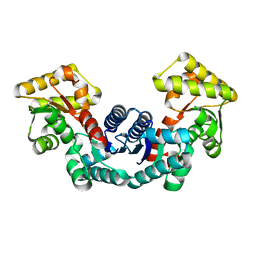 | |
5ELM
 
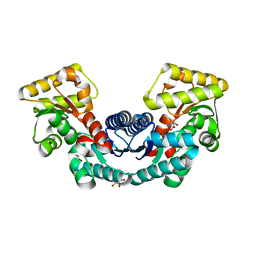 | | Crystal structure of L-aspartate/glutamate specific racemase in complex with L-glutamate | | Descriptor: | Asp/Glu_racemase family protein, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Ahn, J.W, Chang, J.H, Kim, K.J. | | Deposit date: | 2015-11-04 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for an atypical active site of an l-aspartate/glutamate-specific racemase from Escherichia coli
Febs Lett., 589, 2015
|
|
4PZC
 
 | |
4PZE
 
 | |
4PZD
 
 | |
2OPR
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
