6EUG
 
 | |
5G2U
 
 | | Structure of BT1596,a 2-O GAG sulfatase | | Descriptor: | 2-O GLYCOSAMINOGLYCAN SULFATASE, CITRIC ACID, ZINC ION | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2V
 
 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2T
 
 | | BT1596 in complex with its substrate 4,5 unsaturated uronic acid alpha 1,4 D-Glucosamine-2-N, 6-O-disulfate | | Descriptor: | 1,2-ETHANEDIOL, 2-O GLYCOSAMINOGLYCAN SULFATASE, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8R73
 
 | | Polysaccharide lyase BcPL33HA (BcellWH2_04512) Apo form | | Descriptor: | Heparinase II/III-like protein, ZINC ION | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R6Z
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Apo form 2 | | Descriptor: | Heparinase, ZINC ION | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R75
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Apo form 1 | | Descriptor: | Heparinase, ZINC ION | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R70
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HA dp4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparinase, ... | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R72
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HA dp4 collected at 1.33 A | | Descriptor: | Heparinase, ZINC ION, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R71
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HAdp4 collected at 1.22 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparinase, ZINC ION, ... | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
3QEE
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | ACETATE ION, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QEF
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QED
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, CALCIUM ION, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
6EUI
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUJ
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUF
 
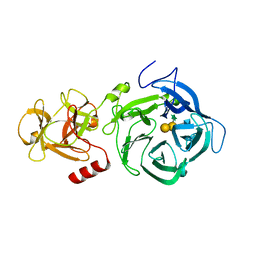 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase, alpha-L-arabinofuranose-(1-3)-[alpha-L-arabinofuranose-(1-4)][beta-D-glucopyranuronic acid-(1-6)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose, alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-6)-[alpha-L-arabinofuranose-(1-3)][alpha-L-arabinofuranose-(1-4)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUH
 
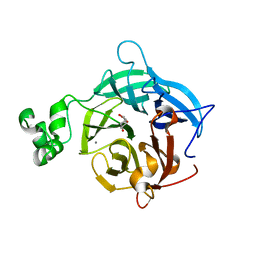 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-glucanase, CALCIUM ION | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
7OZC
 
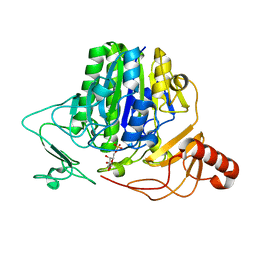 | |
7OZ8
 
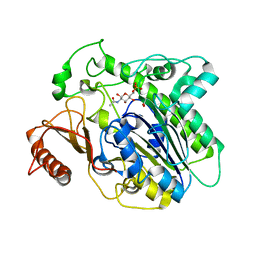 | |
7P26
 
 | |
7P24
 
 | |
7OZA
 
 | |
7OZ9
 
 | |
7OZE
 
 | |
2VX4
 
 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C NATIVE FORM | | Descriptor: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, GLYCEROL, SODIUM ION | | Authors: | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
