6EM9
 
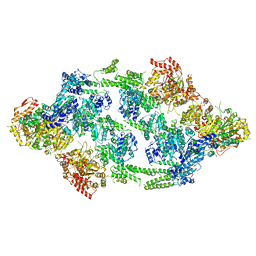 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EMW
 
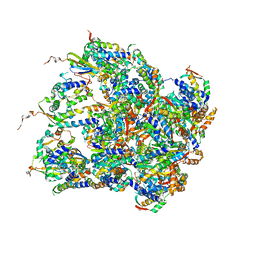 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EM8
 
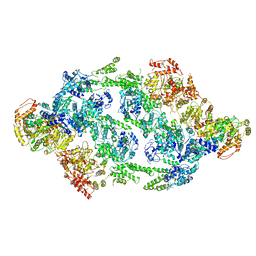 | | S.aureus ClpC resting state, C2 symmetrised | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
4YZS
 
 | |
4YZY
 
 | |
4D2U
 
 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2Q
 
 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
7QVK
 
 | | NM-02 in complex with HER2-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NM-02, Receptor tyrosine-protein kinase erbB-2, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Co-crystallisation and humanisation of an anti-HER2 single-domain antibody as a theranostic tool.
Plos One, 18, 2023
|
|
7BNV
 
 | | Crystal Structure of the SARS-CoV-2 Receptor Binding Domain in Complex with Antibody ION-300 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain, Light Chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M. | | Deposit date: | 2021-01-22 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses.
Front Immunol, 12, 2021
|
|
8BB7
 
 | |
8B3K
 
 | |
8BF4
 
 | | Crystal structure of Mouse Plexin-B1 (20-535) in complex with VHH15 and VHH14 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nanobody inhibitors of Plexin-B1 identify allostery in plexin-semaphorin interactions and signaling.
J.Biol.Chem., 299, 2023
|
|
6SS4
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS2
 
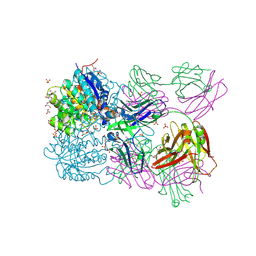 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS0
 
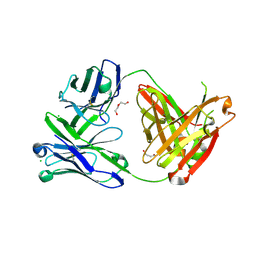 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS6
 
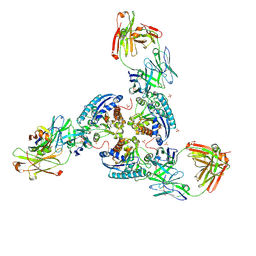 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | Arginase-2, mitochondrial, Fab C0020187 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SRX
 
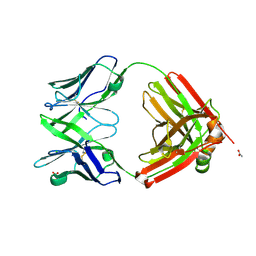 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SRV
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6TUL
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS5
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7NP1
 
 | | Crystal Structure of the SARS-CoV-2 Receptor Binding Domain in Complex with Antibody ION-360 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin light chain, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses.
Front Immunol, 12, 2021
|
|
1POS
 
 | | CRYSTAL STRUCTURE OF A NOVEL DISULFIDE-LINKED "TREFOIL" MOTIF FOUND IN A LARGE FAMILY OF PUTATIVE GROWTH FACTORS | | Descriptor: | PORCINE PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | De, A, Brown, D, Gorman, M, Carr, M, Sanderson, M.R, Freemont, P.S. | | Deposit date: | 1993-10-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a disulfide-linked "trefoil" motif found in a large family of putative growth factors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
5BVN
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-[5-({[(3-fluorophenyl)carbamoyl]amino}methyl)-2-methylphenyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVW
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
