1Q6X
 
 | | Crystal structure of rat choline acetyltransferase | | Descriptor: | SODIUM ION, choline O-acetyltransferase | | Authors: | Cai, Y, Rodgers, D.W. | | Deposit date: | 2003-08-14 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Choline acetyltransferase structure reveals distribution of mutations that cause motor disorders.
Embo J., 23, 2004
|
|
4ZKI
 
 | | The crystal structure of Histidine Kinase YycG with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase | | Authors: | Cai, Y. | | Deposit date: | 2015-04-30 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U7N
 
 | | Inactive structure of histidine kinase | | Descriptor: | Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U7O
 
 | | Active histidine kinase bound with ATP | | Descriptor: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7BQK
 
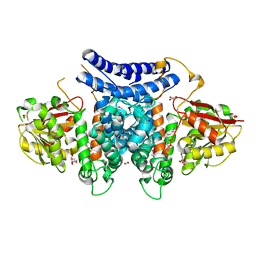 | | The structure of PdxI in complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQL
 
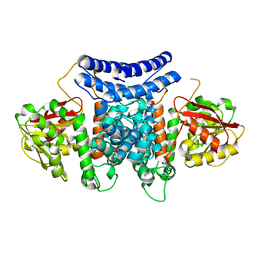 | | The crystal structure of PdxI complex with the Alder-ene adduct | | Descriptor: | 3-[(1R,2S,4R,6S)-2-ethenyl-4,6-dimethyl-cyclohexyl]-4-oxidanyl-1H-pyridin-2-one, DI(HYDROXYETHYL)ETHER, Methyltransf_2 domain-containing protein | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQJ
 
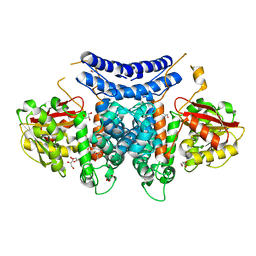 | | The structure of PdxI | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyltransf_2 domain-containing protein, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQO
 
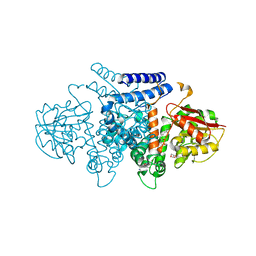 | | The structure of HpiI in complex with its substrate analogue | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-25 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQP
 
 | | The structure of HpiI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-25 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
3CUE
 
 | | Crystal structure of a TRAPP subassembly activating the Rab Ypt1p | | Descriptor: | GTP-binding protein YPT1, PALMITIC ACID, Transport protein particle 18 kDa subunit, ... | | Authors: | Cai, Y, Reinisch, K.M. | | Deposit date: | 2008-04-16 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes.
Cell(Cambridge,Mass.), 133, 2008
|
|
6N5V
 
 | | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine | | Descriptor: | 2-(1H-indol-4-yl)ethan-1-amine, Strictosidine synthase | | Authors: | Cai, Y, Shao, N, Xie, H, Futamura, Y, Panjikar, S, Liu, H, Zhu, H, Osada, H, Zou, H. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine
to be published
|
|
8GNJ
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GNI
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
4TU3
 
 | | Crystal structure of yeast Sac1/Vps74 complex | | Descriptor: | Phosphoinositide phosphatase SAC1, Vacuolar protein sorting-associated protein 74 | | Authors: | Cai, Y, Horenkamp, F.A, Reinisch, K.R. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Sac1-Vps74 structure reveals a mechanism to terminate phosphoinositide signaling in the Golgi apparatus.
J.Cell Biol., 206, 2014
|
|
8GQ5
 
 | |
5C93
 
 | | Histidine kinase with ATP | | Descriptor: | Histidine kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SULFATE ION | | Authors: | Cai, Y. | | Deposit date: | 2015-06-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5CVV
 
 | | coniferyl alcohol bound monolignol 4-O-methyltransferase 9 | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cai, Y, Liu, C.-J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of coniferyl alcohol bound monolignol 4-O-methyltransferase 9 at 1.73 Angstroms resolution
To Be Published
|
|
4I5S
 
 | | Structure and function of sensor histidine kinase | | Descriptor: | Putative histidine kinase CovS; VicK-like protein | | Authors: | Cai, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains
Plos Biol., 11, 2013
|
|
5CVU
 
 | | sinpyl alcohol bound monolignol 4-O-methyltransferase 5 | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, NITRATE ION, ... | | Authors: | Cai, Y, Liu, C.-J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of sinapyl alcohol bound monolignol 4-O-methyltransferase at 1.60 Angstroms resolution
To Be Published
|
|
5CVJ
 
 | | Monolignol 4-O-methyltransferase 5 - coniferyl alcohol | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, NITRATE ION, ... | | Authors: | Cai, Y, Liu, C.-J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of coniferyl alcohol bound monolignol 4-O-methyltransferase at 1.68 Angstroms resolution
To Be Published
|
|
5Z0Q
 
 | | Crystal Structure of OvoB | | Descriptor: | Aminotransferase, class I and II, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cai, Y.J, Huang, P, Wu, L, Zhou, J.H, Liu, P.H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C-S Lyase and Methyltransferase Reactions.
Org. Lett., 20, 2018
|
|
5ZA3
 
 | |
6IX8
 
 | | The structure of LepI C52A in complex with SAM and its substrate analogue | | Descriptor: | (1R,2R,4aS,8S,8aR)-2,8-dimethyl-5'-phenyl-4a,5,6,7,8,8a-hexahydro-2H,2'H-spiro[naphthalene-1,3'-pyridine]-2',4'(1'H)-dione, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX5
 
 | | The structure of LepI complex with SAM and its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX3
 
 | | The structure of LepI complex with SAM | | Descriptor: | CHLORIDE ION, O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
