5MQG
 
 | |
5MPK
 
 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MGE
 
 | |
5MWG
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ091 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-2,3-bis(oxidanylidene)-7-piperidin-1-yl-quinoxalin-6-yl]-5,6,7,8-tetrahydronaphthalene-2-sulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5MGM
 
 | |
5MMG
 
 | | Crystal structure of CREBBP bromodomain complexed with UT07C | | Descriptor: | 1-[4-ethoxy-3-[(1-methylsulfonylindol-6-yl)amino]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MWZ
 
 | |
5MWH
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ089 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-7-morpholin-4-yl-2,3-bis(oxidanylidene)quinoxalin-6-yl]-4-(2-methylpropyl)benzenesulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
6FQT
 
 | | Crystal structure of CREBBP bromodomain complexd with DR46 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexd with DR46
To Be Published
|
|
6YKI
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_092 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-ethyl-2-[(2~{S},5~{R})-5-methyl-2-phenyl-morpholin-4-yl]ethanamine | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YKE
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_038 | | Descriptor: | (2~{R})-2-(3-fluorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YKZ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_234 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-1,4,5,6-tetrahydrocyclopenta[c]pyrazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNK
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_068 | | Descriptor: | 6-[[furan-2-ylmethyl(methyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNN
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_135 | | Descriptor: | 6-[[(2-chloranyl-6-fluoranyl-phenyl)methyl-methyl-amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6YOQ
 
 | |
6YNJ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_046 | | Descriptor: | (2~{R})-2-(2-chlorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_096 | | Descriptor: | 1-[(2~{R},5~{S})-2-(3-chlorophenyl)-5-methyl-morpholin-4-yl]-3-methoxy-propan-2-ol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6Y4G
 
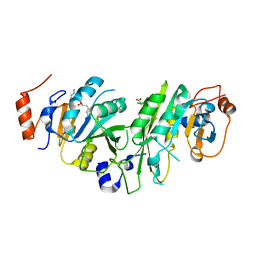 | | Crystal structure of the human METTL3-METTL14 complex bound to Sinefungin | | Descriptor: | ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit, ... | | Authors: | Bedi, R.K, Omori, E, Caflisch, A. | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6YNP
 
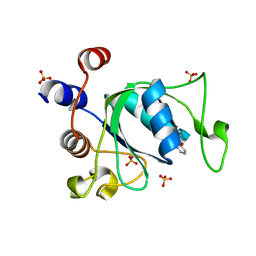 | | Crystal structure of YTHDC1 with compound T96C1 | | Descriptor: | CHLORIDE ION, SULFATE ION, YTHDC1, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YL8
 
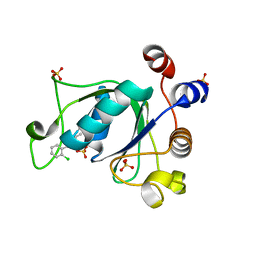 | | Crystal structure of YTHDC1 with compound DHU_DC1_011 | | Descriptor: | (2~{R},5~{S})-2-(2-chlorophenyl)-5-methyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNO
 
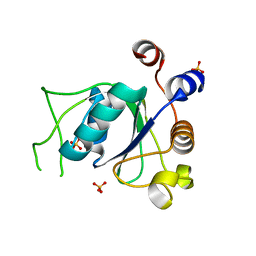 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
7O0M
 
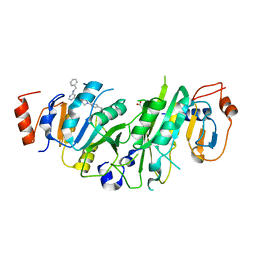 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 9 (ADO_AD_023) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-[6-[(phenylmethyl)amino]pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 9 (ADO_AD_023)
To Be Published
|
|
7O2H
 
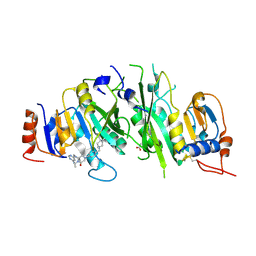 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 13 (ADO_AD_091) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-1-methyl-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 13 (ADO_AD_091)
To Be Published
|
|
7O0R
 
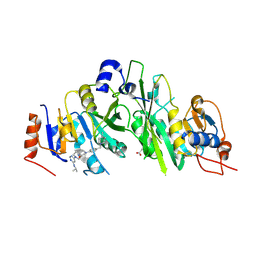 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 15 (ADO_AE_026) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 15 (ADO_AE_026)
To Be Published
|
|
7O27
 
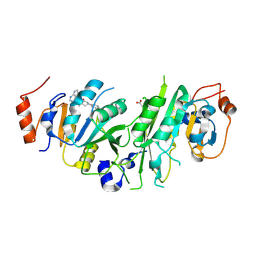 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 17 (ADO_AE_005) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 17 (ADO_AE_005)
To Be Published
|
|
