7DP8
 
 | | Crystal structure of T2R-TTL-Cevipabulin-eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-12-18 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
7EPM
 
 | | human LDHC complexed with NAD+ and ethylamino acetic acid | | Descriptor: | 2-(ethylamino)-2-oxidanylidene-ethanoic acid, L-lactate dehydrogenase C chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-04-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of human LDHC4 as a potential target for anticancer drug discovery.
Acta Pharm Sin B, 12, 2022
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C8K
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGY
 
 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7CR6
 
 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CR8
 
 | | Synechocystis Cas1-Cas2-prespacerL complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7EI1
 
 | | Structure of Pyrococcus furiosus Cas1Cas2 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-03-30 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A distinct structure of Cas1-Cas2 complex provides insights into the mechanism for the longer spacer acquisition in Pyrococcus furiosus.
Int.J.Biol.Macromol., 183, 2021
|
|
4Z8U
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:-ORF2 WITH ATP | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8Q
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 COMPLEX, SELENOMETHIONINE SUBSTITUTED. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8T
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 WITH SULPHATE IONS | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8V
 
 | | CRYSTAL STRUCTURE OF AVRRXO1-ORF1:-ORF2 COMPLEX, NATIVE. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
7P0X
 
 | | Crystal structure of Thioredoxin reductase from Brugia Malayi | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Cheng, Q, Arner, E.S.J, Williams, D.L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
7PVJ
 
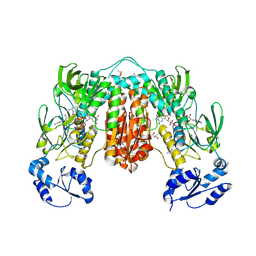 | | Crystal structure of Thioredoxin Reductase from Brugia Malayi in complex with auranofin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Ippoliti, R, Gencheva, R, Cheng, Q, Arner, E.S.J, Angelucci, F, Williams, D.L. | | Deposit date: | 2021-10-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
7PUT
 
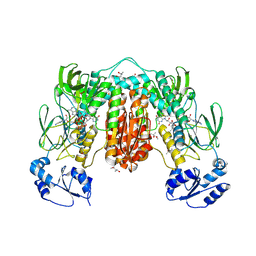 | | Crystal structure of Thioredoxin Reductase from Brugia Malayi in complex with NADP(H) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Ippoliti, R, Gencheva, R, Cheng, Q, Arner, E.S.J, Angelucci, F, Williams, D.L. | | Deposit date: | 2021-09-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
6EDR
 
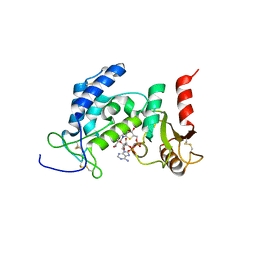 | | Crystal Structure of Human CD38 in Complex with 4'-Thioribose NAD+ | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)thiolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Dai, Z, Zhang, X.N, Nasertorabi, F, Cheng, Q, Pei, H, Louie, S.G, Stevens, C.R, Zhang, Y. | | Deposit date: | 2018-08-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Facile chemoenzymatic synthesis of a novel stable mimic of NAD.
Chem Sci, 9, 2018
|
|
7DCY
 
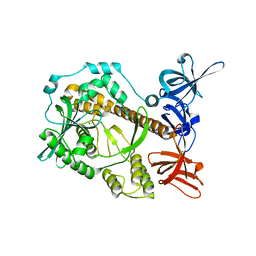 | | Apo form of Mycoplasma genitalium RNase R | | Descriptor: | MAGNESIUM ION, Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DIC
 
 | | Mycoplasma genitalium RNase R in complex with single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DOL
 
 | | Mycoplasma genitalium RNase R in complex with double-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DID
 
 | | Mycoplasma genitalium RNase R in complex with ribose methylated single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
3EAO
 
 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with oxidized C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1, ... | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
3EAN
 
 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with reduced C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
