3LCY
 
 | |
8R84
 
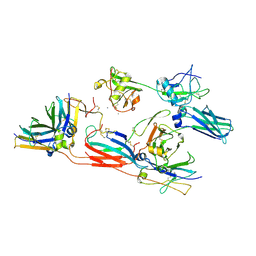 | | pentameric IgMFc-AIM complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CD5 antigen-like, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M.
Nat Commun, 15, 2024
|
|
3LAF
 
 | | Structure of DCC, a netrin-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Deleted in Colorectal Cancer, SULFATE ION, ... | | Authors: | Chen, Q, Liu, J.-H, Wang, J.-H. | | Deposit date: | 2010-01-06 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N-terminal horseshoe conformation of DCC is functionally required for axon guidance and might be shared by other neural receptors.
J.Cell.Sci., 126, 2013
|
|
6D1S
 
 | | Crystal structure of an apo chimeric human alpha1GABAA receptor | | Descriptor: | chimeric alpha1GABAA receptor | | Authors: | Chen, Q, Arjunan, P, Cohen, A.E, Xu, Y, Tang, P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of neurosteroid anesthetic action on GABAAreceptors.
Nat Commun, 9, 2018
|
|
6CNE
 
 | | Selenomethionine variant (V29SeM) of protein GB1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, PHOSPHATE ION | | Authors: | Chen, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CDU
 
 | | Crystal structure of a chimeric human alpha1GABAA receptor in complex with alphaxalone | | Descriptor: | (3a,5a)-3-Hydroxypregnane-11,20-dione, chimeric alpha1GABAA receptor | | Authors: | Chen, Q, Arjunan, P, Cohen, A.E, Xu, Y, Tang, P. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural basis of neurosteroid anesthetic action on GABAAreceptors.
Nat Commun, 9, 2018
|
|
6JV4
 
 | | Crystal structure of metallo-beta-lactamase VMB-1 | | Descriptor: | CITRIC ACID, VMB-1, ZINC ION | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-04-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp.
Adv Biosyst, 4, 2020
|
|
4X9B
 
 | | Crystal structure of Dscam1 isoform 4.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9G
 
 | | Crystal structure of Dscam1 isoform 6.44, N-terminal four Ig domains | | Descriptor: | Down Syndrome Cell Adhesion Molecule isoform 6.44, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X5L
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 9 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AM, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9I
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, isoform 9.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB8
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains (with zinc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4WVR
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9F
 
 | | Crystal structure of Dscam1 isoform 6.9, N-terminal four Ig domains | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Down Syndrome Cell Adhesion Molecule isoform 6.9, GLYCEROL, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X83
 
 | | Crystal structure of Dscam1 isoform 7.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
7MTB
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | Descriptor: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT8
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MTA
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab1 | | Descriptor: | Fab1 Heavy chain, Fab1 Light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT9
 
 | | Rhodopsin kinase (GRK1) in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
6KNZ
 
 | | Crystal structure of T2R-TTL-KXO1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-(2-morpholin-4-ylethoxy)phenyl]pyridin-2-yl]-~{N}-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Reversible binding of the anticancer drug KXO1 (tirbanibulin) to the colchicine-binding site of beta-tubulin explains KXO1's low clinical toxicity.
J.Biol.Chem., 294, 2019
|
|
8R83
 
 | | pentameric IgMFc-AIM complex global refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M.
Nat Commun, 15, 2024
|
|
6J2B
 
 | | CTX-M-64 beta-lactamase S130T sulbactam complex | | Descriptor: | Beta-lactamase, GLYCEROL, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-12-31 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2O
 
 | | Crystal structure of CTX-M-64 clavulanic acid complex | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6ITY
 
 | | CTX-M-64 sulbactam complex | | Descriptor: | ACRYLIC ACID, Beta-lactamase, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
