4HND
 
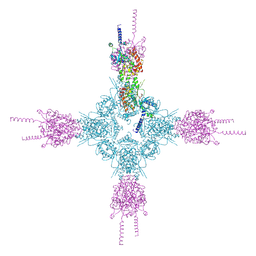 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
4EJW
 
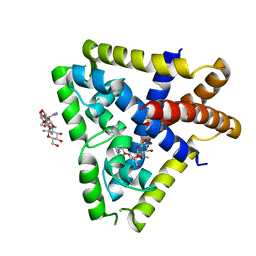 | |
3KVP
 
 | | Crystal Structure of Uncharacterized protein ymzC Precursor from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR378A | | Descriptor: | ACETIC ACID, Uncharacterized protein ymzC | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Afonine, P, Fang, F, Xiao, R, Cunningham, K, Ma, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR378A
To be Published
|
|
7EXH
 
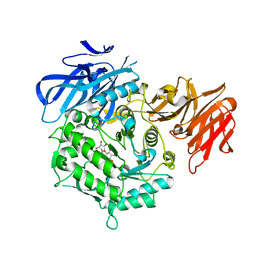 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactinol. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, galactinol | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXR
 
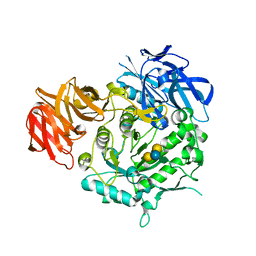 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with Stachyose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXJ
 
 | | Crystal structure of alkaline alpha-galctosidase D383A mutant from Arabidopsis thaliana complexed with Raffinose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXG
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXQ
 
 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with product-galactose and sucrose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5DIB
 
 | | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
4HNE
 
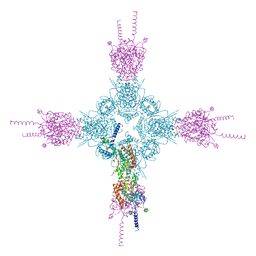 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
2GJ5
 
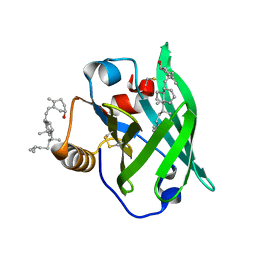 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
3MZ8
 
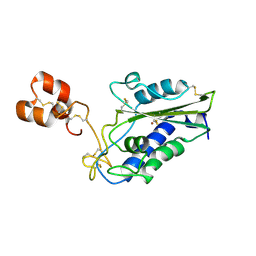 | | Crystal Structure of Zinc-Bound Natrin From Naja atra | | Descriptor: | Natrin-1, ZINC ION | | Authors: | Wang, Y.L, Hsieh, Y.C, Liu, J.S, Chen, C.J, Wu, W.G. | | Deposit date: | 2010-05-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cobra CRISP functions as an inflammatory modulator via a novel Zn2+- and heparan sulfate- dependent transcriptional regulation of endothelial cell adhesion molecules
J.Biol.Chem., 285, 2010
|
|
5WAH
 
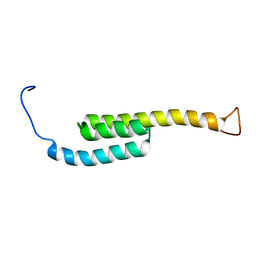 | | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN | | Descriptor: | IgA FC receptor | | Authors: | ELETSKY, A, CHEN, C, FONG, J.J, NIZET, V, VARKI, A, PRESTEGARD, J.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN
To Be Published
|
|
6MUI
 
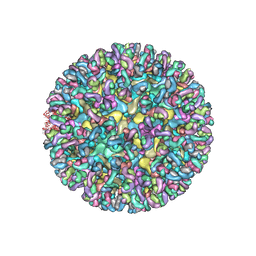 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-42 antibody | | Descriptor: | E1, E2, EEEV-42 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MX7
 
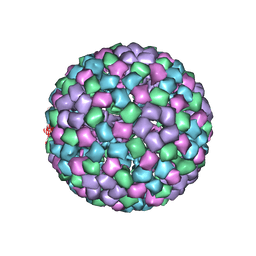 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus: Genome-Binding Capsid N-terminal Domain | | Descriptor: | Capsid | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWV
 
 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-58 Antibody | | Descriptor: | E1, E2, EEEV-58 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MX4
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
5HC6
 
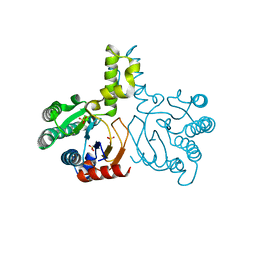 | | Crystal structure of lavandulyl diphosphate synthase from Lavandula x intermedia in apo form | | Descriptor: | SULFATE ION, prenyltransference for protein | | Authors: | Liu, M.X, Liu, W.D, Gao, J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6E2S
 
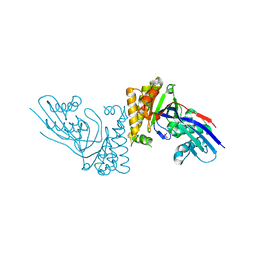 | | apo form of MDDEF with buffer exchange | | Descriptor: | Mevalonate diphosphate decarboxylase, SULFATE ION | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2T
 
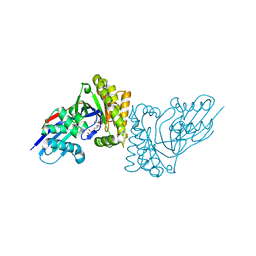 | | MDDEF in complex with MVAPP | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, Mevalonate diphosphate decarboxylase | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2U
 
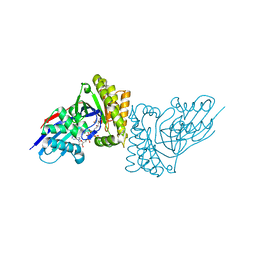 | | MDDEF in complex with MVAPP, AMPPCP and Magnesium | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, MAGNESIUM ION, Mevalonate diphosphate decarboxylase, ... | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2W
 
 | | MDDEF in complex with MVAPP, ADP, sulfate and cobalt | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2V
 
 | | MDDEF in complex with MVAPP, ADPBeF3 and magnesium | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2Y
 
 | | MDDEF in complex with MVAPP, ADP, sulfate and cobalt. Anomalous data | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
