6JCV
 
 | | Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH7.5 | | Descriptor: | MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JCZ
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADPH, and CPD at pH7.5 | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6IQC
 
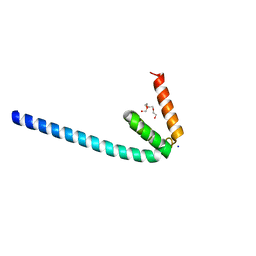 | | Wild-type Programmed Cell Death 5 protein from Sulfolobus solfataricus | | Descriptor: | DNA-binding protein SSO0352, SODIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Chen, C.Y, Lin, K.F, Hsu, C.Y, Tsai, M.J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the programmed cell death 5 protein from Sulfolobus solfataricus.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6IQO
 
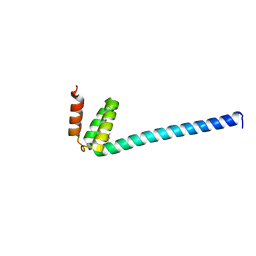 | | Se-Met L45M Programmed Cell Death 5 protein from Sulfolobus solfataricus | | Descriptor: | DNA-binding protein SSO0352, NONAETHYLENE GLYCOL | | Authors: | Chen, C.Y, Lin, K.F, Hsu, C.Y, Tsai, M.J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the programmed cell death 5 protein from Sulfolobus solfataricus.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
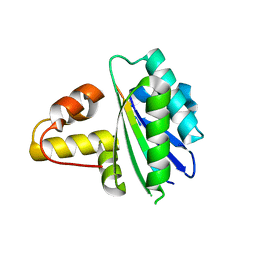
8HDJ
 
 | |
2F43
 
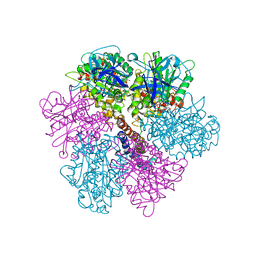 | | Rat liver F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase alpha chain, ... | | Authors: | Chen, C, Saxena, A.K, Simcoke, W.N, Garboczi, D.N, Pedersen, P.L, Ko, Y.H. | | Deposit date: | 2005-11-22 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mitochondrial ATP synthase: Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism.
J.Biol.Chem., 281, 2006
|
|
2GO0
 
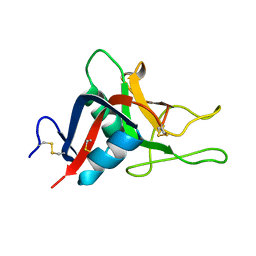 | |
2E8V
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with product GGPP (P21) | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase | | Authors: | Chen, C.K.-M, Guo, R.T, Ko, T.P, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8H61
 
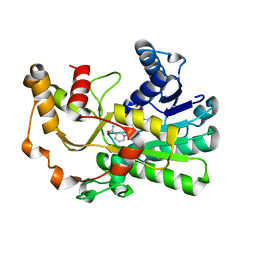 | | Ketoreductase CpKR mutant - M2 | | Descriptor: | Mutant M2 of ketoreductase CpKR, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, C, Pan, J, Xu, J.H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational redesign of a robust ketoreductase for asymmetric synthesis of enantiopure diltiazem precursor.
To Be Published
|
|
3ORF
 
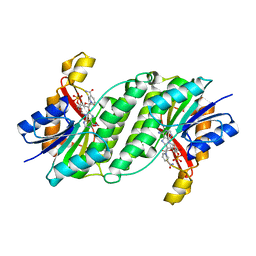 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
2HJP
 
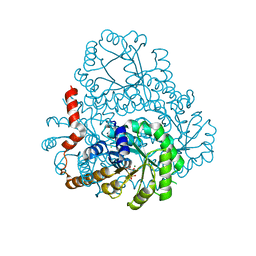 | |
2HRW
 
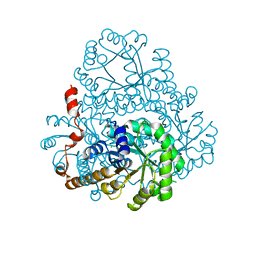 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
4V3Q
 
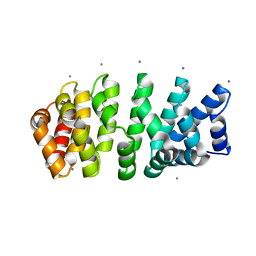 | | Designed armadillo repeat protein with 4 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | CALCIUM ION, GLYCEROL, YIII_M4_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3QYN
 
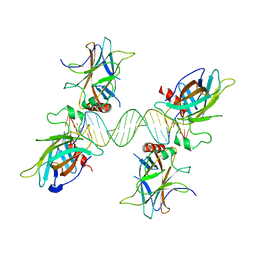 | |
8K32
 
 | |
8W13
 
 | | Crystal structure of MYST acetyltransferase domain in complex with N-(1-(5-bromo-2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)-2-methoxybenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Chen, C, Dou, Y, Wang, M, Xu, C, Buesking, A. | | Deposit date: | 2024-02-15 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification of triazolyl KAT6 inhibitors via a templated fragment approach
Bioorg.Med.Chem.Lett., 2024
|
|
8X3A
 
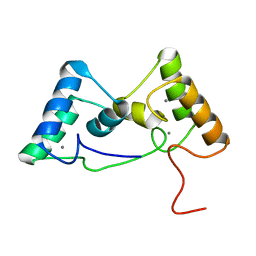 | |
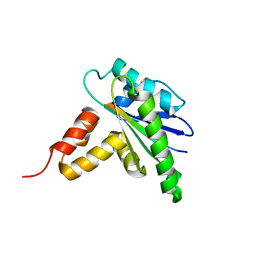
8X39
 
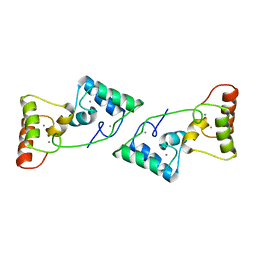 | |
5XB6
 
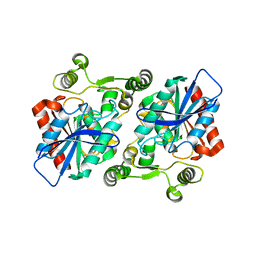 | |
8HER
 
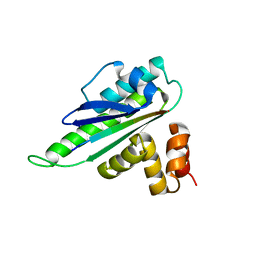 | |
8HEP
 
 | |
8HEQ
 
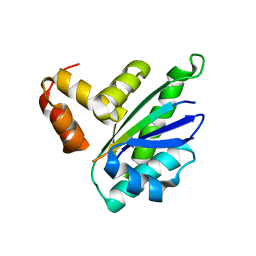 | |
5XKU
 
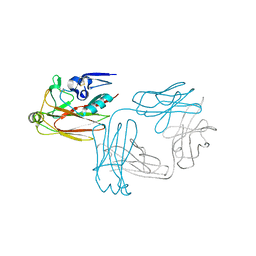 | | Crystal structure of hemagglutinin globular head from an H7N9 influenza virus in complex with a neutralizing antibody HNIgGA6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HNIgGA6 heavy chain, HNIgGA6 light chain, ... | | Authors: | Chen, C, Wang, J, Wang, W, Gao, X, Cui, S, Jin, Q. | | Deposit date: | 2017-05-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Insight into a Human Neutralizing Antibody against Influenza Virus H7N9
J. Virol., 92, 2018
|
|
6A4C
 
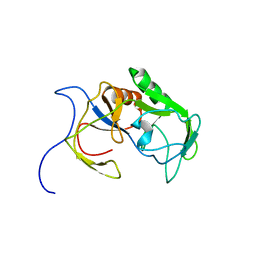 | | Solution structure of MXAN_0049 | | Descriptor: | Uncharacterized protein MXAN_0049 | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MXAN_0049
To Be Published
|
|
6KPE
 
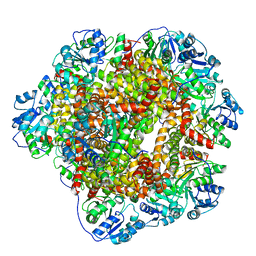 | | 343 K cryoEM structure of Sso-KARI in complex with Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
