3HR5
 
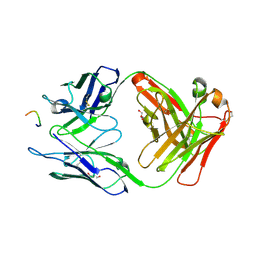 | |
3KOX
 
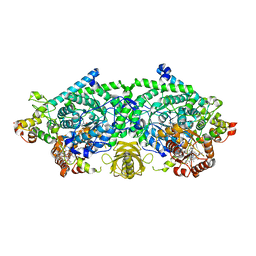 | | Crystal Structure of ornithine 4,5 aminomutase in complex with 2,4-diaminobutyrate (Anaerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KP0
 
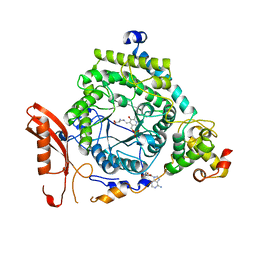 | | Crystal Structure of ORNITHINE 4,5 AMINOMUTASE in complex with 2,4-diaminobutyrate (DAB) (Aerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KFT
 
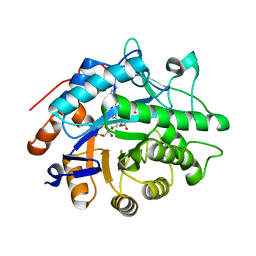 | | Crystal structure of Pentaerythritol Tetranitrate Reductase complex with 1,4,5,6-tetrahydro NADH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Pudney, C.R, Levy, C.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-10-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence to support the hypothesis that promoting vibrations enhance the rate of an enzyme catalyzed H-tunneling reaction.
J.Am.Chem.Soc., 131, 2009
|
|
3KOY
 
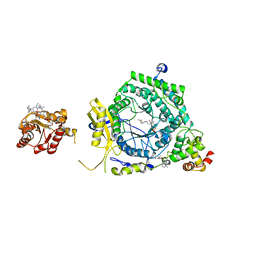 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
1AF2
 
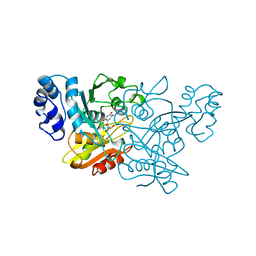 | | CRYSTAL STRUCTURE OF CYTIDINE DEAMINASE COMPLEXED WITH URIDINE | | Descriptor: | CYTIDINE DEAMINASE, URIDINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Xiang, S, Carter, C.W. | | Deposit date: | 1997-03-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization.
Biochemistry, 36, 1997
|
|
1BVO
 
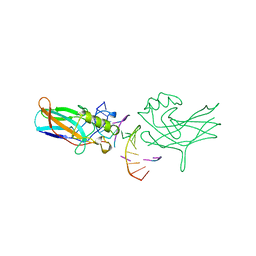 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
1CTU
 
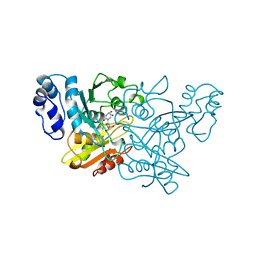 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-ZEBULARINE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
2OTO
 
 | |
1D8A
 
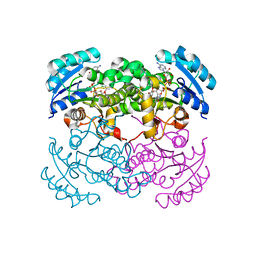 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
1A3Q
 
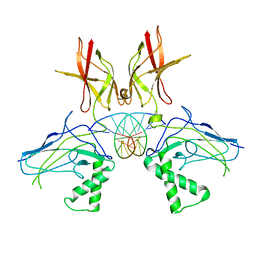 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
1CTT
 
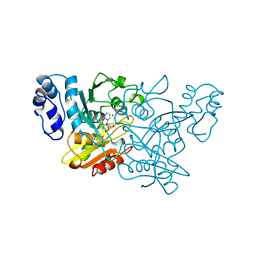 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE NUCLEOSIDE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
1CMY
 
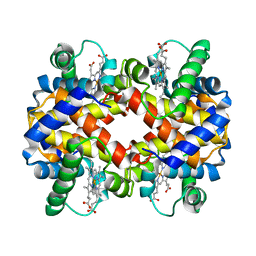 | | THE MUTATION BETA99 ASP-TYR STABILIZES Y-A NEW, COMPOSITE QUATERNARY STATE OF HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN YPSILANTI (CARBONMONOXY) (ALPHA CHAIN), HEMOGLOBIN YPSILANTI (CARBONMONOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smith, F.R, Lattman, E.E, Carter Junior, C.W. | | Deposit date: | 1992-09-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mutation beta 99 Asp-Tyr stabilizes Y--a new, composite quaternary state of human hemoglobin.
Proteins, 10, 1991
|
|
1DKM
 
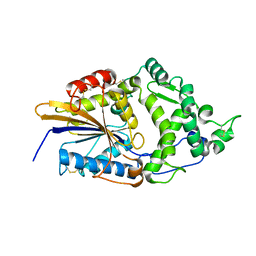 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKQ
 
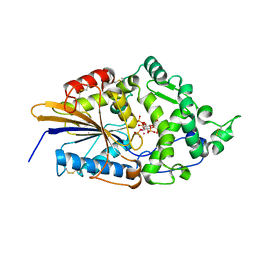 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX ESCHERICHIA COLI PHYTASE AT PH 5.0. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DG4
 
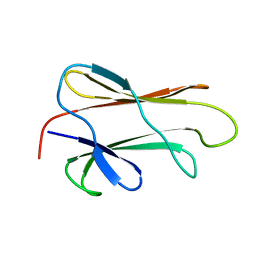 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | Descriptor: | DNAK | | Authors: | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
1ALN
 
 | |
1DKL
 
 | |
1DKO
 
 | | CRYSTAL STRUCTURE OF TUNGSTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH TUNGSTATE BOUND AT THE ACTIVE SITE AND WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE, TUNGSTATE(VI)ION | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKP
 
 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKN
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 5.0 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
5JIX
 
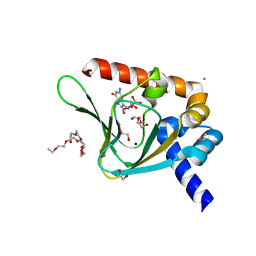 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-Br-cGMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Campbell, J.C, Kim, C.W. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
5JD7
 
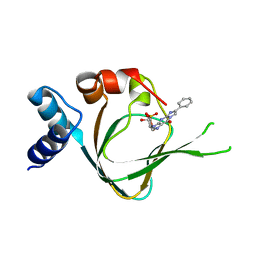 | | PKG I's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with PET-cGMP | | Descriptor: | 3-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-6-phenyl-3,4-dihydro-9H-imidazo[1,2-a]purin-9-one, cGMP-dependent protein kinase 1 | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-04-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
5JAX
 
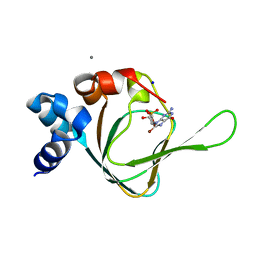 | | PKG I's Carboyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-Br-cGMP | | Descriptor: | 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, SODIUM ION, ... | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
