4IK0
 
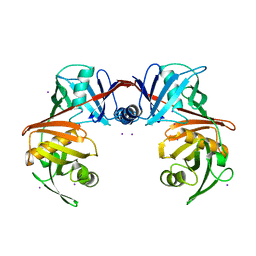 | | Crystal structure of diaminopimelate epimerase Y268A mutant from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, IODIDE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
5WO4
 
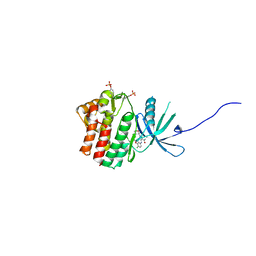 | | JAK1 complexed with compound 28 | | Descriptor: | 3-[(4-chloro-3-methoxyphenyl)amino]-1-[(3R,4S)-4-cyanooxan-3-yl]-1H-pyrazole-4-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lesburg, C.A, Patel, S.B. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Discovery of 3-((4-Chloro-3-methoxyphenyl)amino)-1-((3R,4S)-4-cyanotetrahydro-2H-pyran-3-yl)-1H-pyrazole-4-carboxamide, a Highly Ligand Efficient and Efficacious Janus Kinase 1 Selective Inhibitor with Favorable Pharmacokinetic Properties.
J. Med. Chem., 60, 2017
|
|
2IFU
 
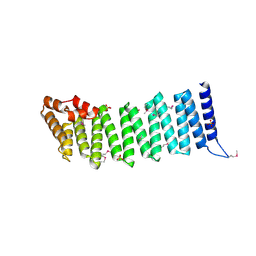 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
6P6L
 
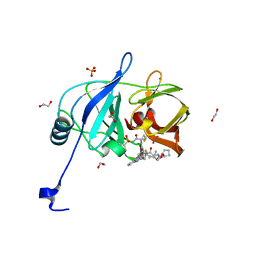 | | HCV NS3/4A protease domain of genotype 1a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.728 Å) | | Cite: | HCV NS3/4A protease domain of genotype 1a in complex with glecaprevir
To Be Published
|
|
6P6S
 
 | | HCV NS3/4A protease domain of genotype 3a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
6P6V
 
 | | HCV NS3/4A protease domain of genotype 5a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
5W2E
 
 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
2IHW
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), apo form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
6P6A
 
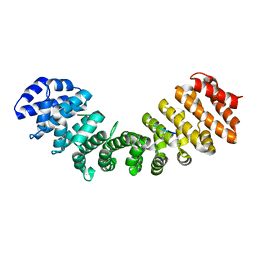 | | Structure of Mouse Importin alpha - NIT2 NLS peptide complex | | Descriptor: | Importin subunit alpha-1, Nitrogen catabolic enzyme regulatory protein | | Authors: | Bernardes, N.E, Fukuda, C.A, Silva, T.D, Oliveira, H.C, Fontes, M.R.M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Comparative study of the interactions between fungal transcription factor nuclear localization sequences with mammalian and fungal importin-alpha.
Sci Rep, 10, 2020
|
|
6P6O
 
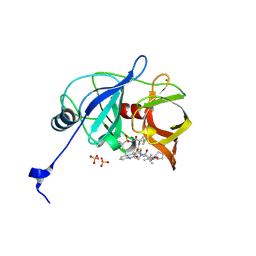 | | HCV NS3/4A protease domain of genotype 1a D168E in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, CHLORIDE ION, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | HCV NS3/4A protease domain of genotype 1a in complex with glecaprevir
To Be Published
|
|
6P6T
 
 | | HCV NS3/4A protease domain of genotype 4a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, Non-structural protein 4A,Serine protease NS3, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
6PJE
 
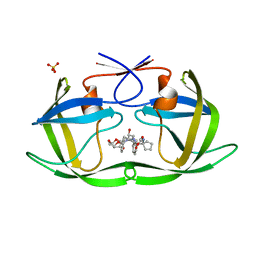 | | HIV-1 Protease NL4-3 WT in Complex with LR2-43 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJO
 
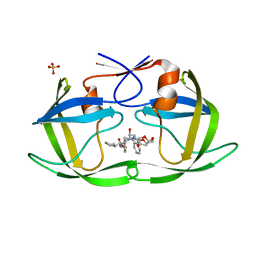 | | HIV-1 Protease NL4-3 WT in Complex with LR2-42 | | Descriptor: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopentyl-2-({(2S,4S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-4-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
4IFW
 
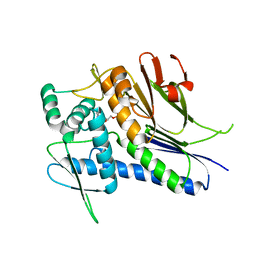 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, ADP inhibited form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
6P99
 
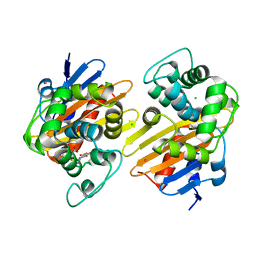 | | OXA-48 carbapanemase, ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6PQC
 
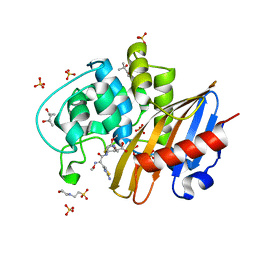 | | Structure of cefotaxime-CDD-1 beta-lactamase complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of CDD-1, the intrinsic class D beta-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime.
J.Struct.Biol., 208, 2019
|
|
2ICX
 
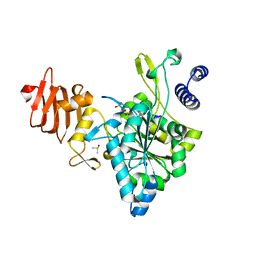 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UTP | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE 5'-TRIPHOSPHATE | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
4IFU
 
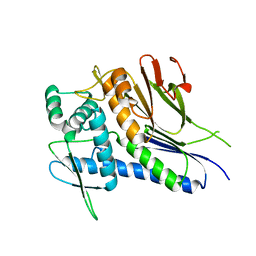 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, apo form | | Descriptor: | FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8334 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
2J1Q
 
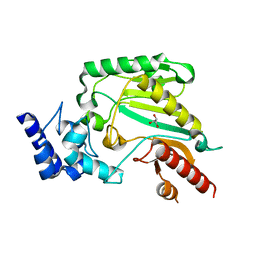 | | Crystal structure of Trypanosoma cruzi arginine kinase | | Descriptor: | ARGININE KINASE, GLYCEROL | | Authors: | Fernandez, P, Haouz, A, Pereira, C.A, Aguilar, C, Alzari, P.M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Arginine Kinase.
Proteins, 69, 2007
|
|
4IHE
 
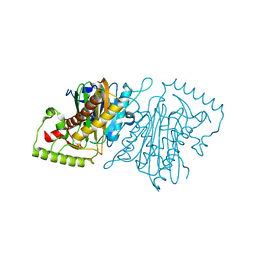 | |
4LG0
 
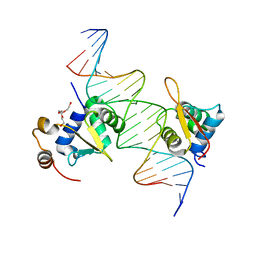 | | Structure of a ternary FOXO1-ETS1 DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DAP*DAP*DCP*DAP*DAP*DTP*DAP*DAP*DCP*DAP*DGP*DGP*DAP*DAP*DAP*DCP*DCP*DGP*DTP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DAP*DCP*DGP*DGP*DTP*DTP*DTP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DTP*DTP*DGP*DT)-3'), ... | | Authors: | Birrane, G, Choy, W.C, Datta, D, Geiger, C.A, Grant, M.A. | | Deposit date: | 2013-06-27 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a ternary FOXO1-ETS1 DNA complex
To be Published
|
|
6PIZ
 
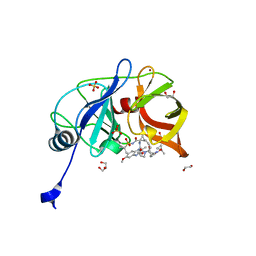 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-1 (NR02-24) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopropyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PQ1
 
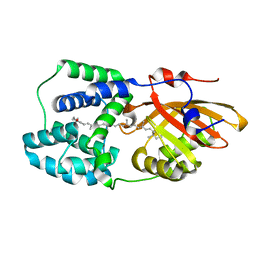 | | Structure of the Fremyella diplosiphon OCP1 | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Dominguez-Martin, M.A, Bao, H, Kerfeld, C.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Comparative ultrafast spectroscopy and structural analysis of OCP1 and OCP2 from Tolypothrix.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
6PJH
 
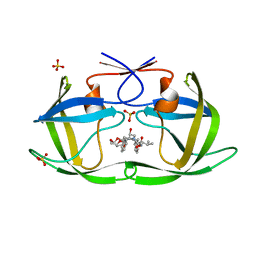 | | HIV-1 Protease NL4-3 WT in Complex with LR3-28 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
