5KVG
 
 | | Zika specific antibody, ZV-67, bound to ZIKA envelope DIII | | Descriptor: | CHLORIDE ION, ZIKA Envelope DIII, ZV-67 Antibody Fab Heavy Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
5KOS
 
 | | Discovery of TAK-272: A Novel, Potent and Orally Active Renin In-hibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-~{tert}-butyl-4-(3-methoxypropylamino)-~{N}-(2-methylpropyl)-~{N}-[(3~{S},5~{R})-5-morpholin-4-ylcarbonylpiperidin-3-yl]pyrimidine-5-carboxamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.-C, Lane, W. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of TAK-272: A Novel, Potent, and Orally Active Renin Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
5KVF
 
 | | Zika specific antibody, ZV-64, bound to ZIKA envelope DIII | | Descriptor: | GLYCEROL, ZV-64 Antibody Fab Heavy Chain, ZV-64 Antibody Fab Light Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
1PSV
 
 | |
5JFL
 
 | |
1LFI
 
 | | METAL SUBSTITUTION IN TRANSFERRINS: THE CRYSTAL STRUCTURE OF HUMAN COPPER-LACTOFERRIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1992-02-10 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metal substitution in transferrins: the crystal structure of human copper-lactoferrin at 2.1-A resolution.
Biochemistry, 31, 1992
|
|
1NQT
 
 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
5KVE
 
 | | Zika specific antibody, ZV-48, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Genome polyprotein, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
1OAH
 
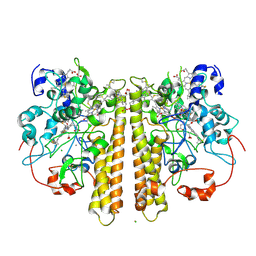 | | Cytochrome c Nitrite Reductase from Desulfovibrio desulfuricans ATCC 27774: The relevance of the two calcium sites in the structure of the catalytic subunit (NrfA). | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Cunha, C.A, Macieira, S, Dias, J.M, Almeida, G, Goncalves, L.L, Costa, C, Lampreia, J, Huber, R, Moura, J.J.G, Moura, I, Romao, M.J. | | Deposit date: | 2003-01-14 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cytochrome C Nitrite Reductase from Desulfovibrio Desulfuricans Atcc 27774. The Relevance of the Two Calcium Sites in the Structure of the Catalytic Subunit (Nrfa)
J.Biol.Chem., 278, 2003
|
|
1QAG
 
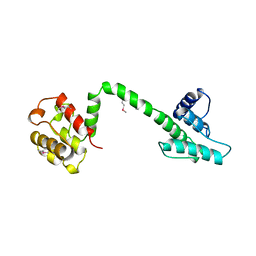 | | Actin binding region of the dystrophin homologue utrophin | | Descriptor: | UTROPHIN ACTIN BINDING REGION | | Authors: | Keep, N.H, Winder, S.J, Moores, C.A, Walke, S, Norwood, F.L.M, Kendrick-Jones, J. | | Deposit date: | 1999-03-05 | | Release date: | 2000-01-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer
Structure Fold.Des., 7, 1999
|
|
1QH8
 
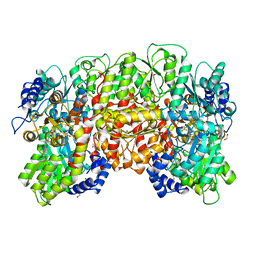 | | NITROGENASE MOFE PROTEIN FROM KLEBSIELLA PNEUMONIAE, AS-CRYSTALLIZED (MIXED OXIDATION) STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
3PI3
 
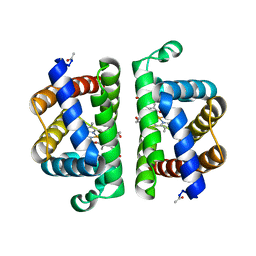 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 5.0 | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
7USZ
 
 | | Human DDAH-1, holo (Zn-bound) form | | Descriptor: | CHLORIDE ION, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, ZINC ION | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UT0
 
 | | Human DDAH-1, apo form | | Descriptor: | 1,2-ETHANEDIOL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UV9
 
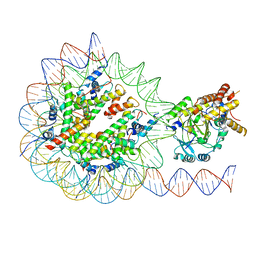 | | KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate | | Descriptor: | DNA (185-MER), FE (III) ION, Histone H2A type 1, ... | | Authors: | Spangler, C.J, Skrajna, A, Foley, C.A, Budziszewski, G.R, Azzam, D.N, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7UVA
 
 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
5KVD
 
 | | Zika specific antibody, ZV-2, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
3PI2
 
 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 8.0 | | Descriptor: | FORMIC ACID, Hemoglobin II, OXYGEN MOLECULE, ... | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
7TVL
 
 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVN
 
 | | Viral AMG chitosanase V-Csn, D148N mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn D148N mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVO
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn E157Q mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
5JU8
 
 | | Cryo-EM structure of an ErmBL-stalled ribosome in complex with P-, and E-tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Bock, L.V, Graf, M, Innis, C.A, Beckmann, R, Grubmueller, H, Vaiana, A.C, Wilson, D.N. | | Deposit date: | 2016-05-10 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest.
Nat Commun, 7, 2016
|
|
5JX4
 
 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
1NR1
 
 | | Crystal structure of the R463A mutant of human Glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
