6NIJ
 
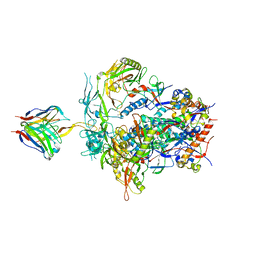 | | PGT145 Fab in complex with full length AMC011 HIV-1 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 Glycoprotein 120, ... | | Authors: | Cottrell, C.A, Torrents de la Pena, A, Rantalainen, K, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-29 | | Release date: | 2019-07-31 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
1JIJ
 
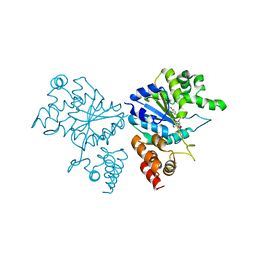 | | Crystal structure of S. aureus TyrRS in complex with SB-239629 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
6OPT
 
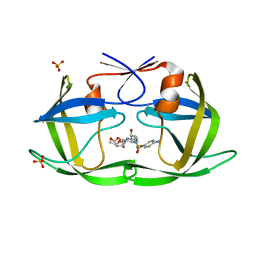 | | HIV-1 Protease NL4-3 V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPX
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
1BQI
 
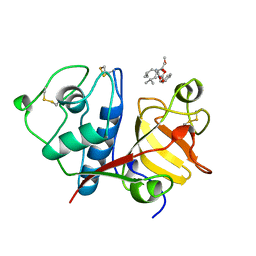 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | Descriptor: | CARBOBENZYLOXY-(L)-LEUCINYL-(L)LEUCINYL METHOXYMETHYLKETONE, PAPAIN | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
1I3J
 
 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF INTRON ENDONUCLEASE I-TEVI WITH ITS SUBSTRATE | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*AP*CP*GP*GP*TP*AP*GP*AP*CP*CP*CP*AP*AP*GP*A)-3', 5'-D(*TP*TP*CP*TP*TP*GP*GP*GP*TP*CP*TP*AP*CP*CP*GP*TP*TP*TP*AP*AP*T)-3', INTRON-ASSOCIATED ENDONUCLEASE 1, ... | | Authors: | Van Roey, P, Waddling, C.A, Fox, K.M, Belfort, M, Derbyshire, V. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate.
EMBO J., 20, 2001
|
|
3O9E
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with af60 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3O9A
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd14 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}propyl]carbamate, PHOSPHATE ION, Pol polyprotein | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
4PTT
 
 | | Crystal Structure of anti-23F strep Fab C05 | | Descriptor: | ACETATE ION, Antibody pn132p2C05, heavy chain, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Smith, K, Schrader, J.W, Pai, E.F. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
6XSU
 
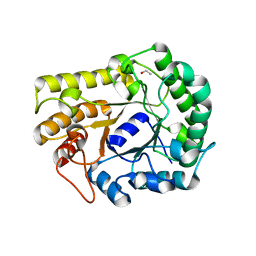 | | GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens | | Descriptor: | ETHANOL, GH5-4 broad specificity endoglucanase | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
3O9D
 
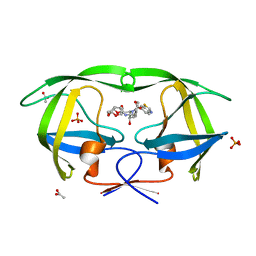 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3O99
 
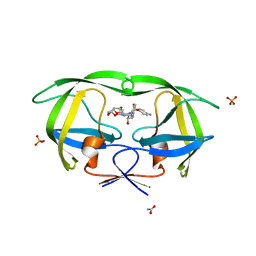 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd13 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
1KF9
 
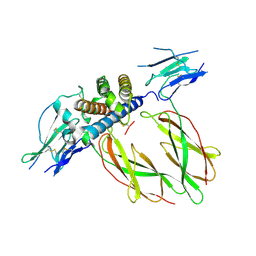 | | PHAGE DISPLAY DERIVED VARIANT OF HUMAN GROWTH HORMONE COMPLEXED WITH TWO COPIES OF THE EXTRACELLULAR DOMAIN OF ITS RECEPTOR | | Descriptor: | EXTRACELLULAR DOMAIN HUMAN GROWTH HORMONE RECEPTOR (1-238), PHAGE DISPLAY DERIVED VARIANT HUMAN GROWTH HORMONE | | Authors: | Schiffer, C.A, Ultsch, M, Walsh, S, Somers, W, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Phage Display Derived Variant of Human Growth Hormone Complexed to Two Copies of the Extracellular Domain of its Receptor: Evidence for Strong Structural Coupling between Receptor Binding Sites
J.Mol.Biol., 316, 2002
|
|
6OPU
 
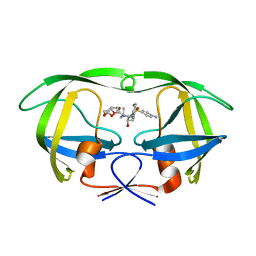 | | HIV-1 Protease NL4-3 K45I, M46I, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPY
 
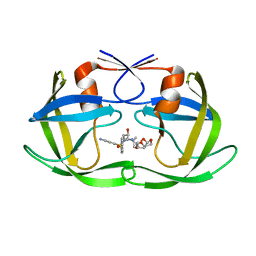 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6XRK
 
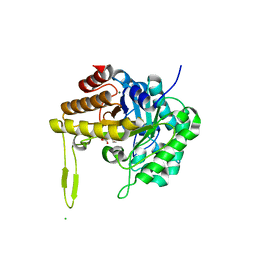 | | GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6XSO
 
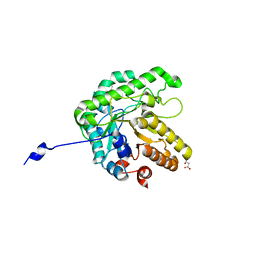 | | GH5-4 broad specificity endoglucanase from an uncultured bacterium | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Cellulase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6OWF
 
 | | Structure of a synthetic beta-carboxysome shell, T=3 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
6O4M
 
 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
4PED
 
 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
1KFS
 
 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*AP*CP*G)-3'), MAGNESIUM ION, PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
6MZU
 
 | |
4Q1Y
 
 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
3Q6S
 
 | |
1JTZ
 
 | | CRYSTAL STRUCTURE OF TRANCE/RANKL CYTOKINE. | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 11 | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity
J.Clin.Invest., 108, 2001
|
|
