2BGD
 
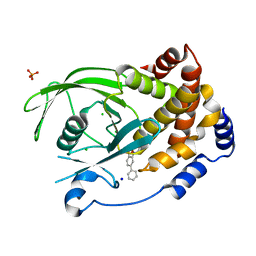 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 5-(4-METHOXYBIPHENYL-3-YL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5V76
 
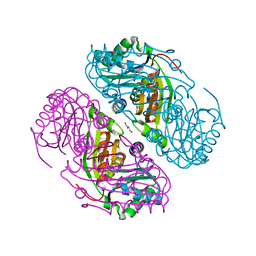 | | Structure of Haliangium ochraceum BMC-T HO-3341 | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Sutter, M, Paasch, B, Zarzycki, J, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell.
Science, 356, 2017
|
|
6PJG
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-97 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-alanyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
4D43
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
5TEI
 
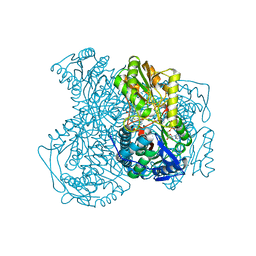 | | Structure of human ALDH1A1 with inhibitor CM039 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-5-(2-methylphenyl)-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ALDH1A1 with inhibitor CM039
To Be Published
|
|
6P97
 
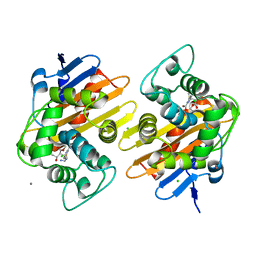 | | OXA-48 carbapanemase, imipenem complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
4D44
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 4-fluoro-2-((2-fluoropyridin-3-yl)oxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5-ethyl-4-fluoro-2-[(2-fluoropyridin-3-yl)oxy]phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
5SZ9
 
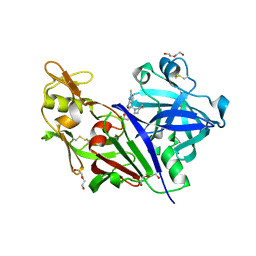 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | (azepan-1-yl)(2-{[(furan-2-yl)methyl]amino}-6-methylpyridin-3-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-08-12 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
2IHW
 
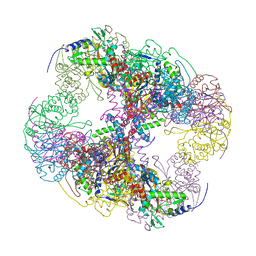 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), apo form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
6MZY
 
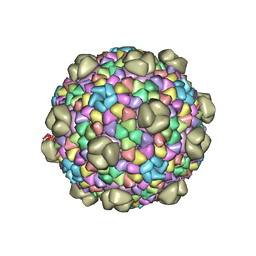 | |
6N0G
 
 | |
6N6Y
 
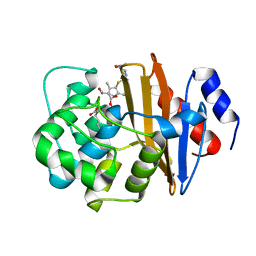 | |
6PIX
 
 | |
2J1Q
 
 | | Crystal structure of Trypanosoma cruzi arginine kinase | | Descriptor: | ARGININE KINASE, GLYCEROL | | Authors: | Fernandez, P, Haouz, A, Pereira, C.A, Aguilar, C, Alzari, P.M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Arginine Kinase.
Proteins, 69, 2007
|
|
6PJC
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR4-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-5-{[(2,6-dimethylphenoxy)acetyl]amino}-4-hydroxy-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.965 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6N6T
 
 | | OXA-23 mutant F110A/M221A low pH form | | Descriptor: | Beta-lactamase oxa23, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D beta-Lactamases.
Antimicrob. Agents Chemother., 63, 2019
|
|
2ICX
 
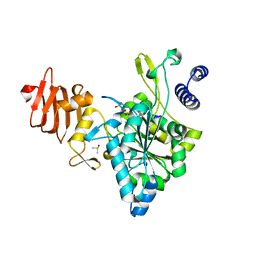 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UTP | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE 5'-TRIPHOSPHATE | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
6MP1
 
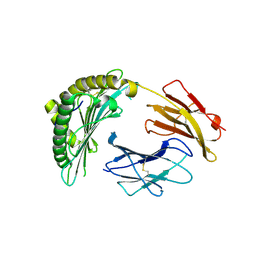 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with the mutant TRP1-K8 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TRP1-K8 peptide, Beta-2-microglobulin,H-2 class I histocompatibility antigen, ... | | Authors: | Clancy-Thompson, E, Devlin, C.A, Birnbaum, M.E, Dougan, S.K. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Altered Binding of Tumor Antigenic Peptides to MHC Class I Affects CD8+T Cell-Effector Responses.
Cancer Immunol Res, 6, 2018
|
|
2IJ0
 
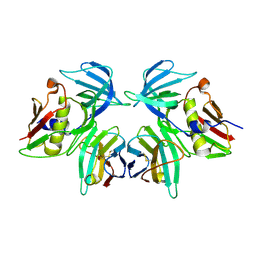 | | Structural basis of T cell specificity and activation by the bacterial superantigen toxic shock syndrome toxin-1 | | Descriptor: | Toxic shock syndrome toxin-1, penultimate affinity-matured variant of hVbeta 2.1, D10 | | Authors: | Moza, B, Varma, A.K, Buonpane, R.A, Zhu, P, Herfst, C.A, Nicholson, M.J, Wilbuer, A.K, Nulifer, S, Wucherpfenning, K.W, McCormick, J.K, Kranz, D.M, Sundberg, E.J. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of T-cell specificity and activation by the bacterial superantigen TSST-1.
Embo J., 26, 2007
|
|
6NER
 
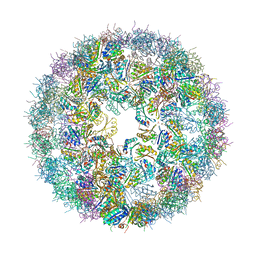 | | Synthetic Haliangium ochraceum BMC shell | | Descriptor: | BMC-H tandem fusion protein, SULFATE ION | | Authors: | Sutter, M, McGuire, S, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies.
ACS Synth Biol, 8, 2019
|
|
2K3V
 
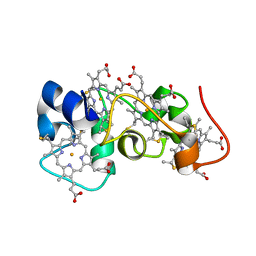 | | Solution Structure of a Tetrahaem Cytochrome from Shewanella Frigidimarina | | Descriptor: | HEME C, Tetraheme cytochrome c-type | | Authors: | Paixao, V.B, Turner, D.L, Salgueiro, C.A, Brennan, L, Reid, G.A, Chapman, S.K. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-31 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a tetraheme cytochrome from Shewanella frigidimarina reveals a novel family structural motif
Biochemistry, 47, 2008
|
|
6PYR
 
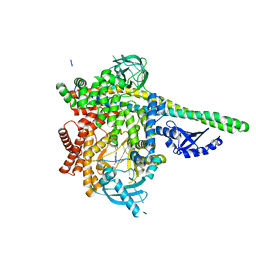 | | Human PI3Kdelta in complex with Compound 2-10 ((3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one) | | Descriptor: | (3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6Q2R
 
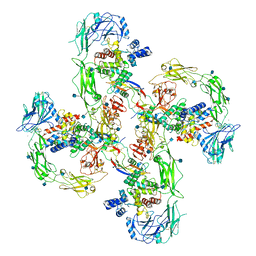 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex in the tetrameric form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
5T39
 
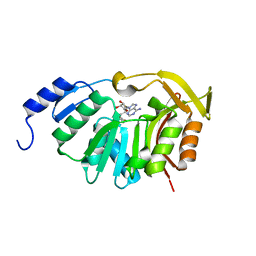 | | Crystal Structure of the N-terminal domain of EvdMO1 in the presence of SAH and D-fucose | | Descriptor: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCulloch, K.M, Starbird, C.A, Chen, Q, Perry, N.A, Berndt, S, Yamakawa, I, Loukachevitch, L.V, Iverson, T.M. | | Deposit date: | 2016-08-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1004 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
5SY2
 
 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, N-ethyl-4-{[(furan-2-yl)methyl]amino}-2-methyl-N-[(3S)-piperidin-3-yl]pyrimidine-5-carboxamide, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
