1SV0
 
 | | Crystal Structure Of Yan-SAM/Mae-SAM Complex | | Descriptor: | Ets DNA-binding protein pokkuri, modulator of the activity of Ets CG15085-PA | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
1VJ3
 
 | | STRUCTURAL STUDIES ON BIO-ACTIVE COMPOUNDS. CRYSTAL STRUCTURE AND MOLECULAR MODELING STUDIES ON THE PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COFACTOR COMPLEX WITH TAB, A HIGHLY SELECTIVE ANTIFOLATE. | | Descriptor: | ACETIC ACID N-[2-CHLORO-5-[6-ETHYL-2,4-DIAMINO-PYRIMID-5-YL]-PHENYL]-[BENZYL-TRIAZEN-3-YL]ETHYL ESTER, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J.R, Queener, S.F, Laughton, C.A, Malcolm, F.G. | | Deposit date: | 2004-01-13 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies on bioactive compounds. 30. Crystal structure and molecular modeling studies on the Pneumocystis carinii dihydrofolate reductase cofactor complex with TAB, a highly selective antifolate.
Biochemistry, 39, 2000
|
|
1THN
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
3TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1 TETRAMUTANT, P2(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
1TID
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: Poised for phosphorylation complex with ATP, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1VY7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VY6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VK5
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, SULFATE ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VJI
 
 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
1VY5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the post-catalysis state of peptide bond formation containing dipeptydil-tRNA in the A site and deacylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VM9
 
 | | The X-ray Structure of the cys84ala cys85ala double mutant of the [2Fe-2S] Ferredoxin subunit of Toluene-4-Monooxygenase from Pseudomonas Mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Moe, L.A, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-13 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of T4moC, the Rieske-type ferredoxin component of toluene 4-monooxygenase.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1NA2
 
 | |
1NLJ
 
 | |
1VTT
 
 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
1VY4
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1W3K
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOBIO DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1W3L
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOTRI DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
3MDF
 
 | | Crystal structure of the RRM domain of Cyclophilin 33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Hom, R.A, Chang, P.Y, Roy, S, Mussleman, C.A, Glass, K.C, Seleznevia, A.I, Gozani, O, Ismagilov, R.F, Cleary, M.L, Kutateladze, T.G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
J.Mol.Biol., 400, 2010
|
|
1W9I
 
 | | Myosin II Dictyostelium discoideum motor domain S456Y bound with MgADP-BeFx | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
3MG2
 
 | | Crystal structure of the orange carotenoid protein Y44S mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
1WA3
 
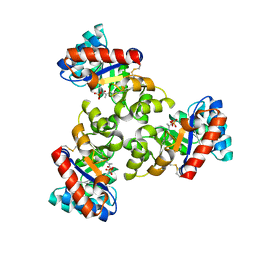 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1W3J
 
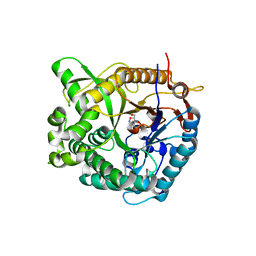 | | Family 1 b-glucosidase from Thermotoga maritima in complex with tetrahydrooxazine | | Descriptor: | BETA-GLUCOSIDASE, TETRAHYDROOXAZINE | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1W9K
 
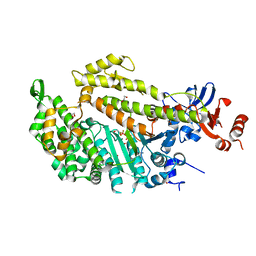 | | Dictyostelium discoideum Myosin II motor domain S456E with bound MgADP-BeFx | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
1NP4
 
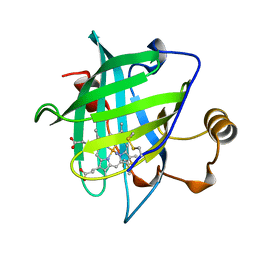 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, PROTEIN (NITROPHORIN 4), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Weichsel, A, Champagne, D.E, Balfour, C.A, Montfort, W.R. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein.
Structure, 6, 1998
|
|
1W9J
 
 | | Myosin II Dictyostelium discoideum motor domain S456Y bound with MgADP-AlF4 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
