2VVD
 
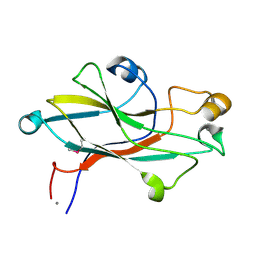 | | Crystal structure of the receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
1SY4
 
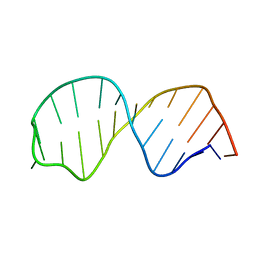 | | Refined solution structure of the S. cerevisiae U6 INTRAMOLECULAR STEM LOOP (ISL) RNA USING RESIDUAL DIPOLAR COUPLINGS (RDCS) | | Descriptor: | U6 Intramolecular Stem-Loop RNA | | Authors: | Reiter, N.J, Nikstad, L.J, Allman, A.M, Johnson, R.J, Butcher, S.E. | | Deposit date: | 2004-03-31 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the U6 RNA intramolecular stem-loop harboring an
S(P)-phosphorothioate modification.
RNA, 9, 2003
|
|
2N7M
 
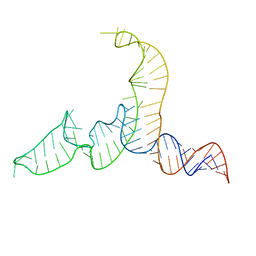 | | NMR-SAXS/WAXS Structure of the core of the U4/U6 di-snRNA | | Descriptor: | RNA (92-MER) | | Authors: | Cornilescu, G, Didychuk, A.L, Rodgers, M.L, Michael, L.A, Burke, J.E, Montemayor, E.J, Hoskins, A.A, Butcher, S.E, Tonelli, M. | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA.
J.Mol.Biol., 428, 2016
|
|
2O32
 
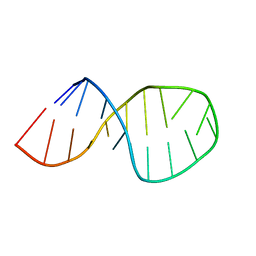 | | Solution structure of U2 snRNA stem I from human, containing modified nucleotides | | Descriptor: | 5'-R(*(PSU)P*CP*UP*CP*(OMG)P*(OMG)P*CP*CP*(PSU)P*UP*UP*UP*(OMG)P*GP*CP*UP*AP*AP*(OMG)P*A)-3' | | Authors: | Sashital, D.G, Venditti, V, Angers, C.G, Cornilescu, G, Butcher, S.E. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human.
Rna, 13, 2007
|
|
1LC6
 
 | | Solution Structure of the U6 Intramolecular Stem-loop RNA | | Descriptor: | U6 Intramolecular Stem-loop RNA | | Authors: | Huppler, A, Nikstad, L.J, Allmann, A.M, Brow, D.A, Butcher, S.E. | | Deposit date: | 2002-04-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure.
Nat.Struct.Biol., 9, 2002
|
|
1SYZ
 
 | |
2KEZ
 
 | | NMR structure of U6 ISL at pH 8.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
2KF0
 
 | | NMR structure of U6 ISL at pH 7.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
1W8X
 
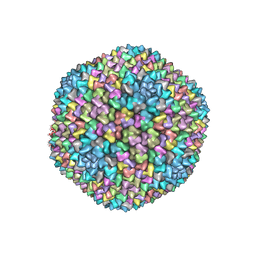 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
2GO9
 
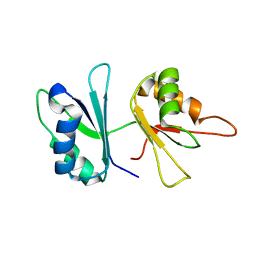 | | RRM domains 1 and 2 of Prp24 from S. cerevisiae | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Reiter, N.J, Lee, D.H, Tonelli, M, Kwan, S.K, Brow, D.A, Butcher, S.E. | | Deposit date: | 2006-04-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
2KH9
 
 | |
1R2P
 
 | | Solution structure of domain 5 from the ai5(gamma) group II intron | | Descriptor: | 34-MER | | Authors: | Sigel, R.K.O, Sashital, D.G, Abramovitz, D.L, Palmer III, A.G, Butcher, S.E, Pyle, A.M. | | Deposit date: | 2003-09-29 | | Release date: | 2004-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2L9W
 
 | |
2L94
 
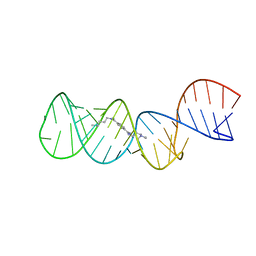 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
2JWV
 
 | |
2I7Z
 
 | |
2I7E
 
 | |
2LK3
 
 | | U2/U6 Helix I | | Descriptor: | RNA (5'-R(*GP*GP*CP*UP*UP*AP*GP*AP*UP*CP*AP*GP*AP*AP*AP*UP*GP*AP*UP*CP*AP*GP*CP*C)-3') | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X.E, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
2LKR
 
 | | Yeast U2/U6 complex | | Descriptor: | RNA (111-MER) | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
2JTP
 
 | |
2JYJ
 
 | | Re-refining the tetraloop-receptor RNA-RNA complex using NMR-derived restraints and Xplor-nih (2.18) | | Descriptor: | RNA (43-MER) | | Authors: | Zuo, X, Wang, J, Foster, T.R, Schwieters, C.D, Tiede, D.M, Butcher, S.E, Wang, Y. | | Deposit date: | 2007-12-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex.
J.Mol.Biol., 351, 2005
|
|
2JUK
 
 | | guanidino neomycin B recognition of an HIV-1 RNA helix | | Descriptor: | (1S,2R,3S,4R,6S)-4,6-bis{[amino(iminio)methyl]amino}-2-{[3-O-(2,6-bis{[amino(iminio)methyl]amino}-2,6-dideoxy-beta-L-glucopyranosyl)-beta-D-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2,6-bis{[amino(iminio)methyl]amino}-2,6-dideoxy-beta-L-glucopyranoside, HIV-1 frameshift site RNA | | Authors: | Staple, D.W, Venditti, V, Niccolai, N, Elson-Schwab, L, Tor, Y, Butcher, S.E. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Guanidinoneomycin B Recognition of an HIV-1 RNA Helix.
Chembiochem, 9, 2008
|
|
