1G7R
 
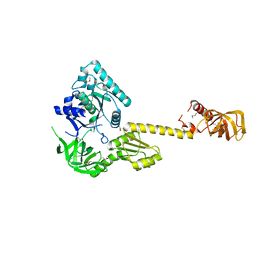 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B | | Descriptor: | TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1EJ4
 
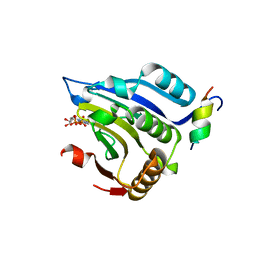 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1G7T
 
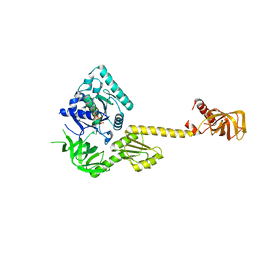 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G62
 
 | | CRYSTAL STRUCTURE OF S.CEREVISIAE EIF6 | | Descriptor: | RIBOSOME ANTI-ASSOCIATION FACTOR EIF6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1DJ8
 
 | |
4F2D
 
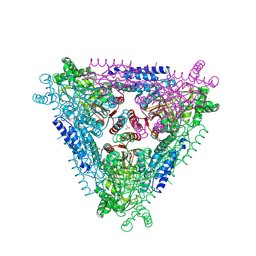 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | Descriptor: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | Authors: | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
4F0S
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine. | | Descriptor: | 5-methylthioadenosine/S-adenosylhomocysteine deaminase, CHLORIDE ION, INOSINE, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine.
To be Published
|
|
4F0R
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
4HL7
 
 | | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae | | Descriptor: | Nicotinate phosphoribosyltransferase, SULFATE ION | | Authors: | Mulichak, A.M, Sauder, J.M, Keefe, L.J, Burley, S.K, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae
To be Published
|
|
4HYR
 
 | | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder
To be published
|
|
4HN8
 
 | | Crystal structure of a putative D-glucarate dehydratase from Pseudomonas mendocina ymp | | Descriptor: | D-glucarate dehydratase, GLYCEROL | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative D-glucarate dehydratase from Pseudomonas mendocina ymp
To be published
|
|
1J2X
 
 | | Crystal structure of RAP74 C-terminal domain complexed with FCP1 C-terminal peptide | | Descriptor: | RNA polymerase II CTD phosphatase, SULFATE ION, Transcription initiation factor IIF, ... | | Authors: | Kamada, K, Roeder, R.G, Burley, S.K. | | Deposit date: | 2003-01-15 | | Release date: | 2003-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of recruitment of TFIIF- associating RNA polymerase C-terminal domain phosphatase (FCP1) by transcription factor IIF
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1ZWK
 
 | |
2A2L
 
 | |
1Q7E
 
 | | Crystal Structure of YfdW protein from E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hypothetical protein yfdW, METHIONINE | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-18 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2A9F
 
 | |
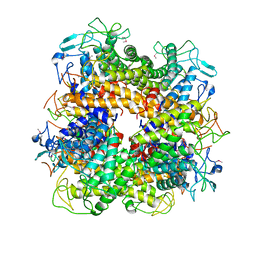
2A1F
 
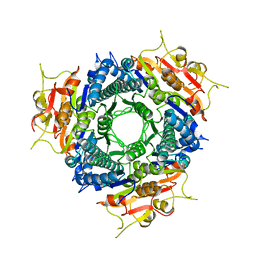 | |
2AA4
 
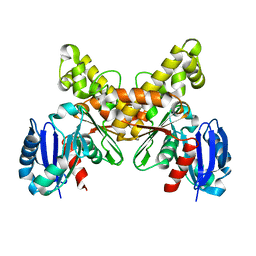 | |
2AFA
 
 | |
2ABQ
 
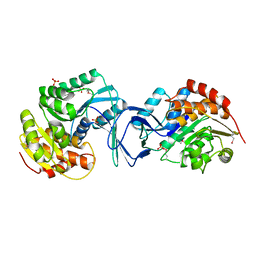 | |
2AKO
 
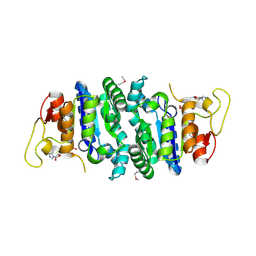 | |
2AP9
 
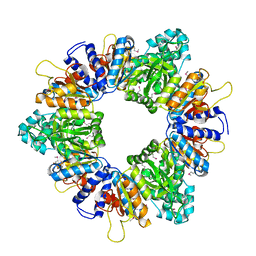 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
1B54
 
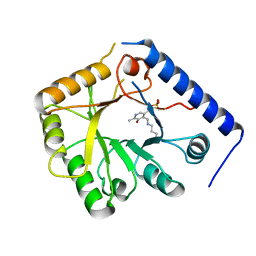 | | CRYSTAL STRUCTURE OF A YEAST HYPOTHETICAL PROTEIN-A STRUCTURE FROM BNL'S HUMAN PROTEOME PROJECT | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, YEAST HYPOTHETICAL PROTEIN | | Authors: | Swaminathan, S, Eswaramoorthy, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-01-12 | | Release date: | 1999-01-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NKP
 
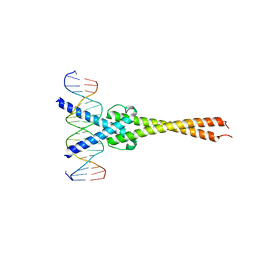 | | Crystal structure of Myc-Max recognizing DNA | | Descriptor: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
3BOX
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg | | Descriptor: | L-rhamnonate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
