1XXL
 
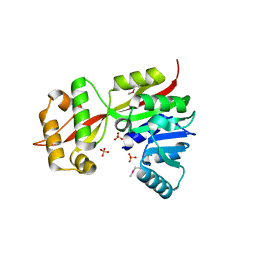 | |
2GPY
 
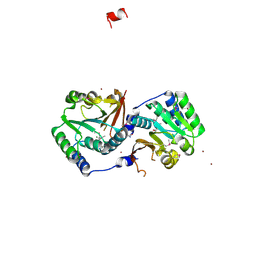 | | Crystal structure of putative O-methyltransferase from Bacillus halodurans | | Descriptor: | MAGNESIUM ION, O-methyltransferase, ZINC ION | | Authors: | Ramagopal, U.A, Patskovsky, Y.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative O-methyltransferase from Bacillus halodurans
To be Published
|
|
2HB6
 
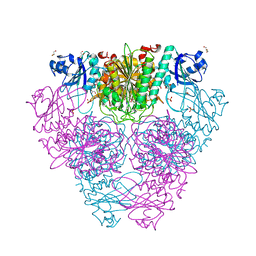 | | Structure of Caenorhabditis elegans leucine aminopeptidase (LAP1) | | Descriptor: | GLYCEROL, Leucine aminopeptidase 1, SODIUM ION, ... | | Authors: | Patskovsky, Y, Zhan, C, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
1Y0G
 
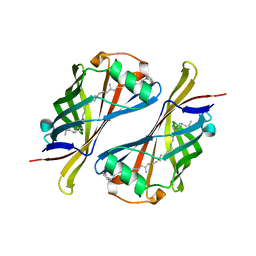 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PROTEIN, STRUCTURAL GENOMICS | | Descriptor: | 2-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL]PHENOL, Protein yceI | | Authors: | Patskovsky, Y.V, Strokopytov, B, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PERIPLASMIC PROTEIN
To be Published
|
|
1Z2I
 
 | |
1Z4E
 
 | |
1YV9
 
 | |
2HJ1
 
 | | Crystal structure of a 3D domain-swapped dimer of protein HI0395 from Haemophilus influenzae | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y.V, Toro, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a 3D domain-swapped dimer of hypothetical protein from Haemophilus influenzae (CASP Target)
To be Published
|
|
3R9K
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asp mutant complexed with sulfate, a closed cap conformation | | Descriptor: | Putative beta-phosphoglucomutase, SULFATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
1YEY
 
 | | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
1YI7
 
 | | Beta-d-xylosidase (selenomethionine) XYND from Clostridium Acetobutylicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-xylosidase from clostridium acetobutylicum
To be Published
|
|
1Y1L
 
 | |
1Y7B
 
 | | BETA-D-XYLOSIDASE, A FAMILY 43 GLYCOSIDE HYDROLASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-08 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of beta-xylosidase from Clostridium acetobutylicum
To be Published
|
|
1Y9E
 
 | |
1YAV
 
 | |
1YBE
 
 | |
1Y7E
 
 | |
1YGA
 
 | |
1Y8C
 
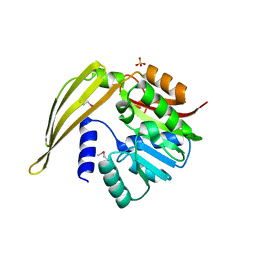 | | Crystal structure of a S-adenosylmethionine-dependent methyltransferase from Clostridium acetobutylicum ATCC 824 | | Descriptor: | S-adenosylmethionine-dependent methyltransferase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-11 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a S-adenosylmethionine-dependent methyltransferase from Clostridium acetobutylicum ATCC 824
To be Published
|
|
1YBD
 
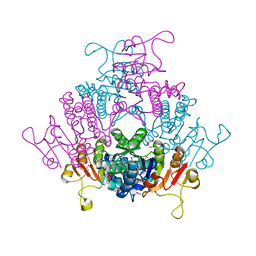 | |
1YS9
 
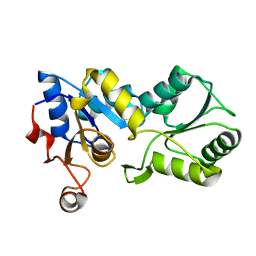 | |
1YRZ
 
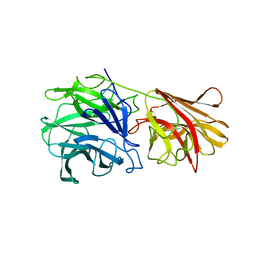 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
1YMY
 
 | | Crystal Structure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the N-acetylglucosamine-6-phosphate deacetylase from Escherichia Coli
To be Published
|
|
3OYR
 
 | | Crystal structure of polyprenyl synthase from Caulobacter crescentus CB15 complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, PYROPHOSPHATE 2-, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2F02
 
 | |
