3GRA
 
 | | Crystal structure of AraC family transcriptional regulator from Pseudomonas putida | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-25 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AraC family transcriptional regulator from Pseudomonas putida
To be Published
|
|
3G7U
 
 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
3GD5
 
 | | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Ramagopal, U.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus
To be Published
|
|
5Q03
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methylpropyl)thiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(2-methylpropyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methylpropyl)thiophen-2-yl]sulfonylurea
To be published
|
|
3GG9
 
 | | CRYSTAL STRUCTURE OF putative D-3-phosphoglycerate dehydrogenase oxidoreductase from Ralstonia solanacearum | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Putative D-3-Phosphoglycerate Dehydrogenase from Ralstonia Solanacearum
To be Published
|
|
5Q01
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-[(3,4-dichloro-1-methyl-1H-pyrazol-5-yl)oxy]benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea
To be published
|
|
3GPI
 
 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
3GRZ
 
 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
5PZR
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[3-[3-[(3-chlorophenyl)sulfonylcarbamoylamino]propoxy]propyl]urea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N,N'-{oxybis[(propane-3,1-diyl)carbamoyl]}bis(3-chlorobenzene-1-sulfonamide) | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[3-[3-[(3-chlorophenyl)sulfonylcarbamoylamino]propoxy]propyl]urea
To be published
|
|
5Q09
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-[4-bromo-6-(carbamoylamino)pyridin-2-yl]-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-{[4-bromo-6-(carbamoylamino)pyridin-2-yl]carbamoyl}-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-[4-bromo-6-(carbamoylamino)pyridin-2-yl]-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea
To be published
|
|
3GMG
 
 | | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv1825/MT1873 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis
To be Published
|
|
3GMS
 
 | |
3H7L
 
 | | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus | | Descriptor: | ENDOGLUCANASE, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-27 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus
To be Published
|
|
3GM8
 
 | | Crystal structure of a beta-glycosidase from Bacteroides vulgatus | | Descriptor: | GLYCEROL, Glycoside hydrolase family 2, candidate beta-glycosidase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a beta-glycosidase from Bacteroides vulgatus
To be Published
|
|
3H75
 
 | | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens | | Descriptor: | GLYCEROL, Periplasmic sugar-binding domain protein, SULFATE ION | | Authors: | Kumaran, D, Mahmood, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens
To be Published
|
|
3HDV
 
 | |
3HEB
 
 | |
3HG9
 
 | | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192 | | Descriptor: | NICKEL (II) ION, PilM | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192
To be Published
|
|
3HDC
 
 | |
5Q04
 
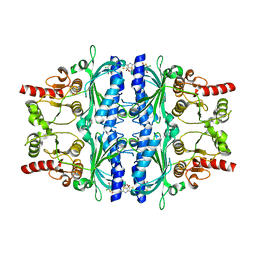 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea | | Descriptor: | 4-bromo-N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-chlorothiophene-2-sulfonamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea
To be published
|
|
3HUH
 
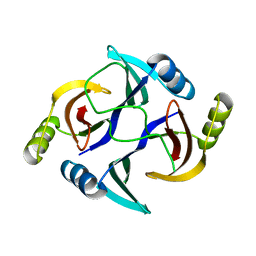 | | The structure of biphenyl-2,3-diol 1,2-dioxygenase iii-related protein from salmonella typhimurium | | Descriptor: | Virulence protein STM3117 | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-14 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of biphenyl-2,3-diol 1,2-dioxygenase iii-related protein from salmonella typhimurium
To be Published
|
|
5Q05
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea
To be published
|
|
3HUR
 
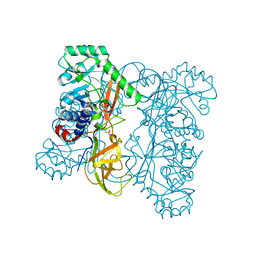 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, SULFATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of alanine racemase from Oenococcus oeni
To be Published
|
|
3JTE
 
 | | Crystal structure of response regulator receiver domain Protein from clostridium thermocellum | | Descriptor: | Response regulator receiver protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of response regulator receiver domain
Protein from clostridium thermocellum
To be Published
|
|
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
