6CBE
 
 | | Atomic structure of a rationally engineered gene delivery vector, AAV2.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Burg, M, Rosebrough, C, Drouin, L, Bennett, A, Mietzsch, M, Chipman, P, McKenna, R, Sousa, D, Potter, M, Byrne, B, Kozyreva, O.G, Samulski, R.J, Agbandje-McKenna, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Atomic structure of a rationally engineered gene delivery vector, AAV2.5.
J. Struct. Biol., 203, 2018
|
|
1MNT
 
 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
7TVW
 
 | | Crystal structure of Arabidopsis thaliana DLK2 | | Descriptor: | Alpha/beta-Hydrolases superfamily protein | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Arabidopsis DWARF14-LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture.
Plant Direct, 6, 2022
|
|
7UOC
 
 | | Crystal structure of Orobanche minor KAI2d4 | | Descriptor: | CHLORIDE ION, KAI2d4 | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Divergent Clade KAI2 Protein in the Root Parasitic Plant Orobanche minor Is a Highly Sensitive Strigolactone Receptor and Is Involved in the Perception of Sesquiterpene Lactones.
Plant Cell.Physiol., 64, 2023
|
|
6AZD
 
 | |
5D0J
 
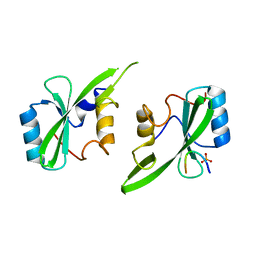 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
6ATX
 
 | |
6AZC
 
 | |
6AZB
 
 | |
6ZVZ
 
 | |
6AVW
 
 | |
6AVY
 
 | |
6AVV
 
 | |
6AVX
 
 | |
1ZNJ
 
 | | INSULIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Turkenburg, J.P, Dodson, G.G, Derewenda, U, Smith, G.D, Dodson, E.J, Derewenda, Z.S, Xiao, B. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Determination and Refinement of Two Crystal Forms of Native Insulins
To be Published
|
|
3U0V
 
 | | Crystal Structure Analysis of human LYPLAL1 | | Descriptor: | Lysophospholipase-like protein 1 | | Authors: | Burger, M, Zimmermann, T.J, Kondoh, Y, Stege, P, Watanabe, N, Osada, H, Waldmann, H, Vetter, I.R. | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases
J.Lipid Res., 53, 2012
|
|
1ZNI
 
 | | INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, ZINC ION | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Dodson, G.G, Dodson, E.J, Xiao, B, Bentley, G.A. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of insulin in 4-zinc insulin.
Nature, 261, 1976
|
|
1ADZ
 
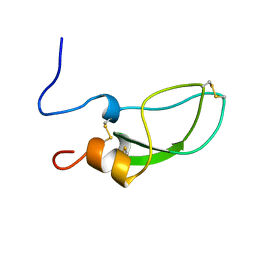 | |
5VYE
 
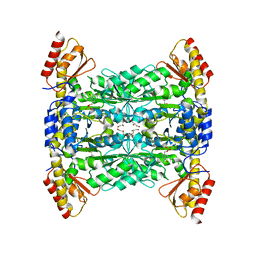 | |
5VG3
 
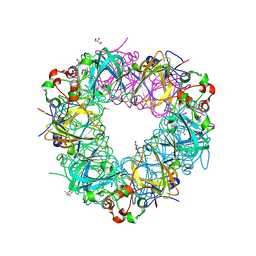 | |
6TZP
 
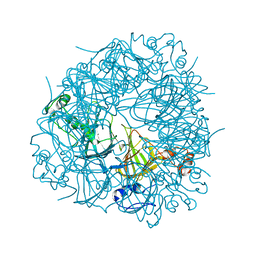 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
6UFI
 
 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
4UZR
 
 | |
4WG7
 
 | | Room-temperature crystal structure of lysozyme determined by serial synchrotron crystallography using a nano focused beam. | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kappe, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WG1
 
 | | Room temperature crystal structure of lysozyme determined by serial synchrotron crystallography (micro focused beam - crystFEL) | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kapp, U, Shilova, A, Weimhausen, B, Sauter, N.K, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-09-17 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
