4UF1
 
 | | Deerpox virus DPV022 in complex with Bak BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2L4F
 
 | |
2L4E
 
 | |
5HUD
 
 | | Non-covalent complex of and DAHP synthase and chorismate mutase from Corynebacterium glutamicum with bound transition state analog | | Descriptor: | 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
1N71
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii in complex with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Ghuman, N, Wybenga-Groot, L.E, Berghuis, A.M. | | Deposit date: | 2002-11-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the AAC(6')-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members
Protein Sci., 12, 2003
|
|
6B15
 
 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K | | Descriptor: | 1,2-ETHANEDIOL, Amy13K | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-16 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
6SS4
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS2
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS0
 
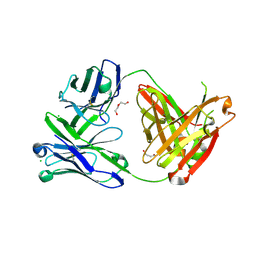 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS6
 
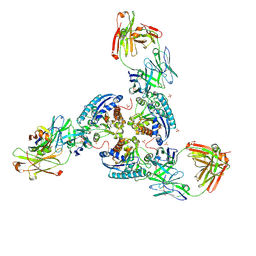 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | Arginase-2, mitochondrial, Fab C0020187 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SRX
 
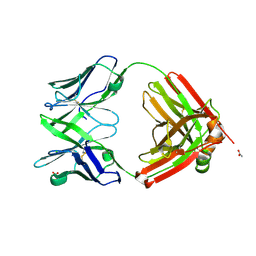 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
4YS7
 
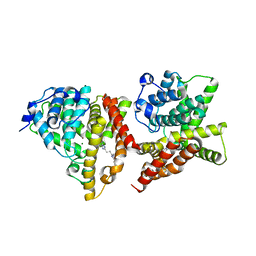 | | Co-crystal structure of 2-[2-(5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl)ethyl]-3-methyl-3H-imidazo[4,5-f]quinoline (compound 39) with PDE10A | | Descriptor: | 2-[2-(5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl)ethyl]-3-methyl-3H-imidazo[4,5-f]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Burdi, D.F, Herman, L, Wang, T. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Evolution and synthesis of novel orally bioavailable inhibitors of PDE10A.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6Z5S
 
 | | RC-LH1(14)-W complex from Rhodopseudomonas palustris | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (6~{R},10~{S},14~{R},19~{R},23~{S},24~{E},27~{S},28~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-24,28-dien-2-ol, ... | | Authors: | Swainsbury, D.J.K, Qian, P, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2020-05-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels.
Sci Adv, 7, 2021
|
|
6Z5R
 
 | | RC-LH1(16) complex from Rhodopseudomonas palustris | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (6~{R},10~{S},14~{R},19~{R},23~{S},24~{E},27~{S},28~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-24,28-dien-2-ol, ... | | Authors: | Swainsbury, D.J.K, Qian, P, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2020-05-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels.
Sci Adv, 7, 2021
|
|
6SRV
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS5
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1OK7
 
 | | A Conserved protein binding-site on Bacterial Sliding Clamps | | Descriptor: | DNA POLYMERASE III, DNA POLYMERASE IV | | Authors: | Burnouf, D.Y, Olieric, V, Wagner, J, Fujii, S, Reinbolt, J, Fuchs, R.P.P, Dumas, P. | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of Sliding Clamp/Ligand Interactions Suggest a Competition between Replicative and Translesion DNA Polymerases
J.Mol.Biol., 335, 2004
|
|
6I82
 
 | | Crystal structure of partially phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018412 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
4YQH
 
 | | 2-[2-(4-Phenyl-1H-imidazol-2-yl)ethyl]quinoxaline (Sunovion Compound 14) co-crystallized with PDE10A | | Descriptor: | 2-[2-(4-phenyl-1H-imidazol-2-yl)ethyl]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Burdi, D, Herman, L, Wang, T. | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Evolution and synthesis of novel orally bioavailable inhibitors of PDE10A.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6TUL
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6I83
 
 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
1RSE
 
 | |
2A4N
 
 | | Crystal structure of aminoglycoside 6'-N-acetyltransferase complexed with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Xiong, B, Breitbach, C, Berghuis, A.M. | | Deposit date: | 2005-06-29 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of aminoglycoside acetyltransferase AAC(6')-Ii in a novel crystal form: structural and normal-mode analyses.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6B3P
 
 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
