2Y3S
 
 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y4G
 
 | | Structure of the Tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P212121 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2011-01-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
1Q3K
 
 | |
2FX8
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with an aib-induced peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41) | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
1NXQ
 
 | | Crystal Structure of R-alcohol dehydrogenase (RADH) (apoenyzme) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-alcohol dehydrogenase | | Authors: | Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2003-02-11 | | Release date: | 2003-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency
J.Mol.Biol., 327, 2003
|
|
1PZ3
 
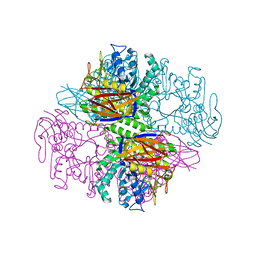 | | Crystal structure of a family 51 (GH51) alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T6 | | Descriptor: | Alpha-L-arabinofuranosidase, GLYCEROL | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1TF8
 
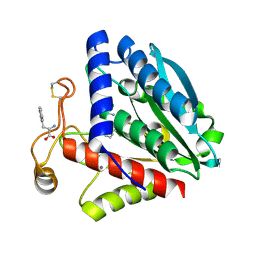 | | Streptomyces griseus aminopeptidase complexed with L-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2KRG
 
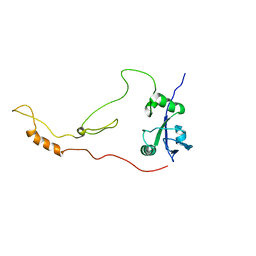 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
2JWH
 
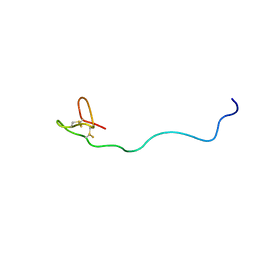 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
1PZ2
 
 | | Crystal structure of a transient covalent reaction intermediate of a family 51 alpha-L-arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
2JWG
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
2NLO
 
 | |
2KJD
 
 | |
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1DXY
 
 | | STRUCTURE OF D-2-HYDROXYISOCAPROATE DEHYDROGENASE | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, D-2-HYDROXYISOCAPROATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dengler, U, Niefind, K, Kiess, M, Schomburg, D. | | Deposit date: | 1996-08-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 A resolution.
J.Mol.Biol., 267, 1997
|
|
2MOV
 
 | | Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal Derived AGEs. | | Descriptor: | Advanced glycosylation end product-specific receptor, N~5~-[(5R)-5-methyl-4-oxo-4,5-dihydro-1H-imidazol-2-yl]-L-ornithine | | Authors: | Shekhtman, A, Xue, J, Ray, R, Singer, D, Bohme, D, Burz, D.S, Rai, V, Hoffman, R. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal-Derived AGEs.
Biochemistry, 53, 2014
|
|
4JY4
 
 | | Crystal structure of human Fab PGT121, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PGT121 heavy chain, ... | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
1ZK2
 
 | | Orthorhombic crystal structure of the apo-form of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic resolution structures of R-specific alcohol dehydrogenase from Lactobacillus brevis provide the structural bases of its substrate and cosubstrate specificity
J.Mol.Biol., 349, 2005
|
|
1ZK0
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1GAL
 
 | | CRYSTAL STRUCTURE OF GLUCOSE OXIDASE FROM ASPERGILLUS NIGER: REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, GLUCOSE OXIDASE, ... | | Authors: | Hecht, H.J, Kalisz, K, Hendle, J, Schmid, R.D, Schomburg, D. | | Deposit date: | 1992-08-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glucose oxidase from Aspergillus niger refined at 2.3 A resolution.
J.Mol.Biol., 229, 1993
|
|
1ZJZ
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NAD | | Descriptor: | (1R)-1-PHENYLETHANOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
1ZK1
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NAD | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZK4
 
 | | Structure of R-specific alcohol dehydrogenase (wildtype) from Lactobacillus brevis in complex with acetophenone and NADP | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZJY
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
