4N48
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with capped RNA fragment | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
8HD7
 
 | | The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The intermediate pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8HD6
 
 | | The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8I7N
 
 | | The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanylpurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD7
 
 | | The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The pre-Tet-C state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD3
 
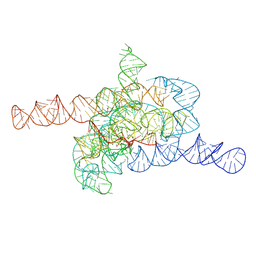 | | The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD4
 
 | | The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | Co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon, MAGNESIUM ION | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD5
 
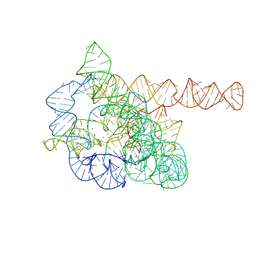 | | The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD6
 
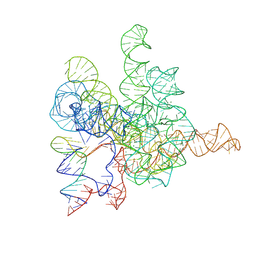 | | The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
6FPT
 
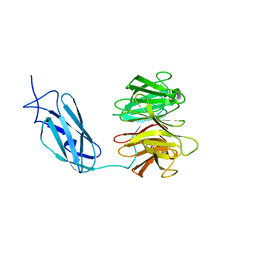 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | Descriptor: | E3 ubiquitin-protein ligase TRIM71 | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQL
 
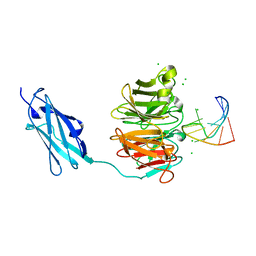 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQ3
 
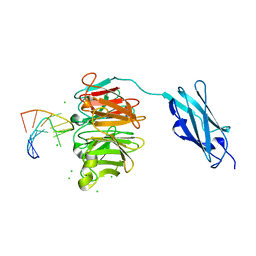 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6G6S
 
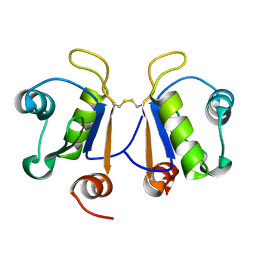 | | Crystal structure of human Acinus RNA recognition motif domain | | Descriptor: | Apoptotic chromatin condensation inducer in the nucleus | | Authors: | Fernandes, H, Czapinska, H, Grudziaz, K, Bujnicki, J.M, Nowacka, M. | | Deposit date: | 2018-04-03 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human Acinus RNA recognition motif domain.
PeerJ, 6, 2018
|
|
6QM2
 
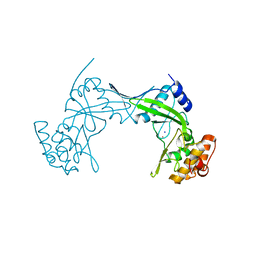 | | NlaIV restriction endonuclease | | Descriptor: | POTASSIUM ION, SODIUM ION, Type-2 restriction enzyme NlaIV | | Authors: | Czapinska, H, Siwek, W, Szczepanowski, R.H, Bujnicki, J.M, Bochtler, M, Skowronek, K. | | Deposit date: | 2019-02-01 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure and Directed Evolution of Specificity of NlaIV Restriction Endonuclease.
J.Mol.Biol., 431, 2019
|
|
6IA2
 
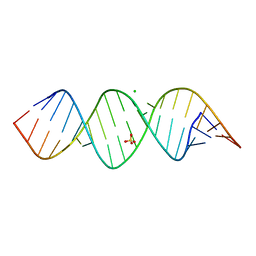 | | Crystal structure of a self-complementary RNA duplex recognized by Com | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*GP*AP*GP*AP*AP*CP*CP*CP*GP*GP*AP*GP*UP*UP*CP*CP*CP*U)-3'), SULFATE ION | | Authors: | Nowacka, M, Fernandes, H, Kiliszek, A, Bernat, A, Lach, G, Bujnicki, J.M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specific interaction of zinc finger protein Com with RNA and the crystal structure of a self-complementary RNA duplex recognized by Com.
Plos One, 14, 2019
|
|
7YR7
 
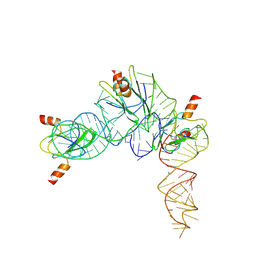 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
8QO4
 
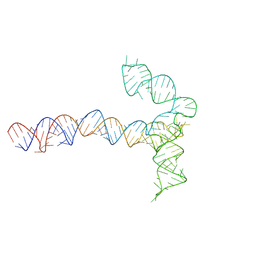 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO3
 
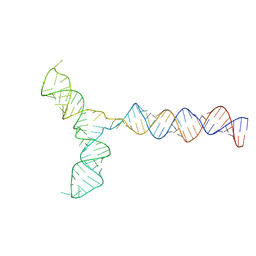 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO5
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO2
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
9G7C
 
 | | Cryo-EM structure of raiA ncRNA from Clostridium reveals a new RNA 3D fold | | Descriptor: | raiA (224-MER) | | Authors: | Badepally, N.G, Moura, T.R, Purta, E, Baulin, E, Bujnicki, J.M. | | Deposit date: | 2024-07-20 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structure of raiA ncRNA from Clostridium reveals a new RNA 3D fold
J.Mol.Biol., 2024
|
|
4KYW
 
 | | Restriction endonuclease DPNI in complex with two DNA molecules | | Descriptor: | 5'-(*DC*DTP*DGP*DGP*6MAP*DTP*DCP*DCP*DAP*DG)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Mierzejewska, K, Siwek, W, Czapinska, H, Skowronek, K, Bujnicki, J.M, Bochtler, M. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the methylation specificity of R.DpnI.
Nucleic Acids Res., 42, 2014
|
|
2Y7H
 
 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
4ESJ
 
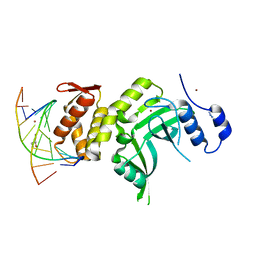 | | RESTRICTION ENDONUCLEASE DpnI IN COMPLEX WITH TARGET DNA | | Descriptor: | AZIDE ION, DNA (5'-D(*CP*TP*GP*GP*(6MA)P*TP*CP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Siwek, W, Czapinska, H, Bochtler, M, Bujnicki, J.M, Skowronek, K. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI.
Nucleic Acids Res., 40, 2012
|
|
2Y7C
 
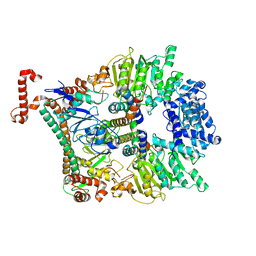 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
