6EE5
 
 | |
2VH4
 
 | | Structure of a loop C-sheet serpin polymer | | 分子名称: | TENGPIN | | 著者 | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | 登録日 | 2007-11-19 | | 公開日 | 2008-01-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
5J7C
 
 | |
5J7K
 
 | |
1YZW
 
 | | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore | | 分子名称: | DI(HYDROXYETHYL)ETHER, GFP-like non-fluorescent chromoprotein | | 著者 | Wilmann, P.G, Petersen, J, Pettikiriarachchi, A, Buckle, A.M, Devenish, R.J, Prescott, M, Rossjohn, J. | | 登録日 | 2005-02-28 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore
J.Mol.Biol., 349, 2005
|
|
5CDZ
 
 | | Crystal structure of conserpin in the latent state | | 分子名称: | Conserpin in the latent state, GLYCEROL | | 著者 | Porebski, B.T, McGowan, S, Keleher, S, Buckle, A.M. | | 登録日 | 2015-07-06 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.449 Å) | | 主引用文献 | Smoothing a rugged protein folding landscape by sequence-based redesign.
Sci Rep, 6, 2016
|
|
5CE0
 
 | |
5CDX
 
 | |
4U3H
 
 | |
4F87
 
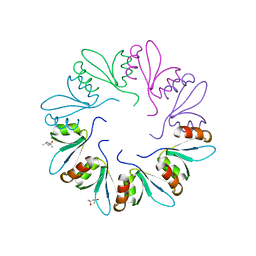 | | X-ray Crystal Structure of PlyCB | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | 著者 | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | 登録日 | 2012-05-17 | | 公開日 | 2012-07-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F88
 
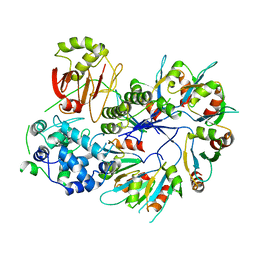 | | X-ray Crystal Structure of PlyC | | 分子名称: | PlyCA, PlyCB | | 著者 | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | 登録日 | 2012-05-17 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1KCQ
 
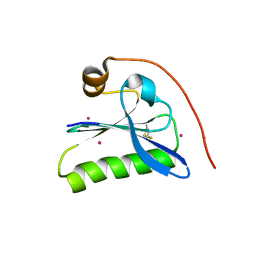 | | Human Gelsolin Domain 2 with a Cd2+ bound | | 分子名称: | CADMIUM ION, GELSOLIN | | 著者 | Kazmirski, S.L, Isaacson, R.L, An, C, Buckle, A, Johnson, C.M, Daggett, V, Fersht, A.R. | | 登録日 | 2001-11-09 | | 公開日 | 2002-01-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.
Nat.Struct.Biol., 9, 2002
|
|
4KGA
 
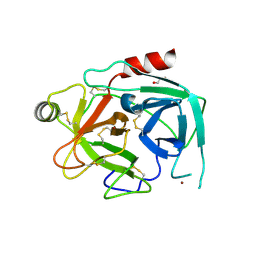 | | Crystal structure of kallikrein-related peptidase 4 | | 分子名称: | 1,2-ETHANEDIOL, Kallikrein-4, NICKEL (II) ION | | 著者 | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | 登録日 | 2013-04-29 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
3JZ4
 
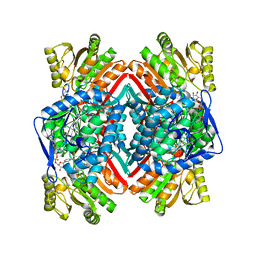 | | Crystal structure of E. coli NADP dependent enzyme | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase [NADP+] | | 著者 | Langendorf, C.G, Key, T.L.G, Fenalti, G, Kan, W.T, Buckle, A.M, Caradoc-Davies, T, Tuck, K.L, Law, R.H.P, Whisstock, J.C. | | 登録日 | 2009-09-22 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions.
Plos One, 5, 2010
|
|
4K1E
 
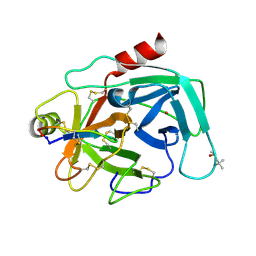 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Kallikrein-4, LITHIUM ION, ... | | 著者 | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | 登録日 | 2013-04-04 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
4KEL
 
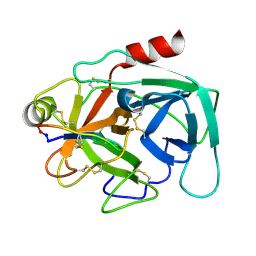 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | 分子名称: | Kallikrein-4, Trypsin inhibitor 1 | | 著者 | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | 登録日 | 2013-04-25 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.148 Å) | | 主引用文献 | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
1SNG
 
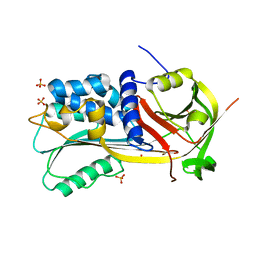 | | Structure of a Thermophilic Serpin in the Native State | | 分子名称: | COG4826: Serine protease inhibitor, SULFATE ION | | 著者 | Fulton, K.F, Buckle, A.M, Cabrita, L.D, Irving, J.A, Butcher, R.E, Smith, I, Reeve, S, Lesk, A.M, Bottomley, S.P, Rossjohn, J, Whisstock, J.C. | | 登録日 | 2004-03-10 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition.
J.Biol.Chem., 280, 2005
|
|
4K8Y
 
 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with Sunflower Trypsin Inhibitor (SFTI-1) | | 分子名称: | Kallikrein-4, Trypsin inhibitor 1 | | 著者 | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | 登録日 | 2013-04-19 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
1WZ9
 
 | | The 2.1 A structure of a tumour suppressing serpin | | 分子名称: | Maspin precursor, SULFATE ION | | 著者 | Law, R.H, Irving, J.A, Buckle, A.M, Ruzyla, K, Buzza, M, Bashtannyk-Puhalovich, T.A, Beddoe, T.C, Kim, N, Worrall, D.M, Bottomley, S.P, Bird, P.I, Rossjohn, J, Whisstock, J.C. | | 登録日 | 2005-03-03 | | 公開日 | 2005-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
1FYA
 
 | |
1FY9
 
 | |
2H4R
 
 | |
2H4Q
 
 | |
3EBG
 
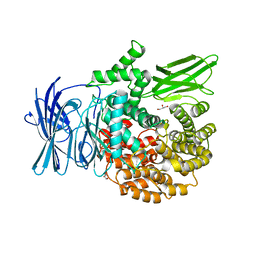 | | Structure of the M1 Alanylaminopeptidase from malaria | | 分子名称: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | 著者 | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | 登録日 | 2008-08-27 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EBH
 
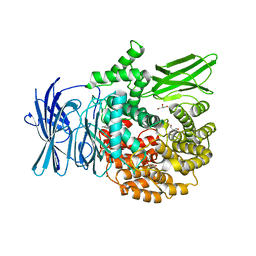 | | Structure of the M1 Alanylaminopeptidase from malaria complexed with bestatin | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | 登録日 | 2008-08-27 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
