1ZKK
 
 | | Crystal structure of hSET8 in ternary complex with H4 peptide (16-24) and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase, H4 lysine-20 specific, Peptide corresponding to residues 15-24 of histone H4, ... | | Authors: | Couture, J.-F, Collazo, E, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
Genes Dev., 19, 2005
|
|
1YSJ
 
 | | Crystal Structure of Bacillus Subtilis YXEP Protein (APC1829), a Dinuclear Metal Binding Peptidase from M20 Family | | Descriptor: | NICKEL (II) ION, protein yxeP | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Bacillus Subtilis YXEP Protein, a Dinuclear Metal Binding Peptidase from M20 Family
To be Published
|
|
1ZSW
 
 | | Crystal Structure of Bacillus cereus Metallo Protein from Glyoxalase family | | Descriptor: | BETA-MERCAPTOETHANOL, Glyoxalase family protein, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Bacillus cereus Metallo Protein from Glyoxalase family
To be Published
|
|
1ZTU
 
 | | Structure of the chromophore binding domain of bacterial phytochrome | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Forest, K.T, Vierstra, R.D. | | Deposit date: | 2005-05-27 | | Release date: | 2005-11-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome.
Nature, 438, 2005
|
|
6D0G
 
 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
6C9H
 
 | | non-phosphorylated AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9J
 
 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Brunzelle, J.S, Hitoshi, Y, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6D57
 
 | | Campylobacter jejuni ferric uptake regulator S1 metalated | | Descriptor: | FORMIC ACID, Ferric uptake regulation protein, GLYCEROL, ... | | Authors: | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | Deposit date: | 2018-04-19 | | Release date: | 2018-05-23 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Functional insights into the interplay between DNA interaction and metal coordination in ferric uptake regulators.
Sci Rep, 8, 2018
|
|
6D0P
 
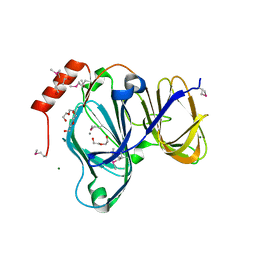 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
6C4V
 
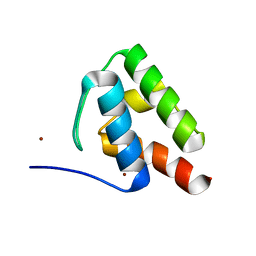 | | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | Polyketide synthase Pks13, ZINC ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To be Published
|
|
6C9F
 
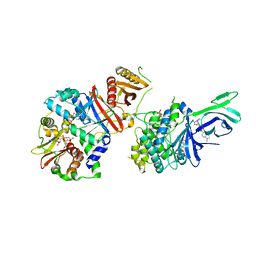 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9G
 
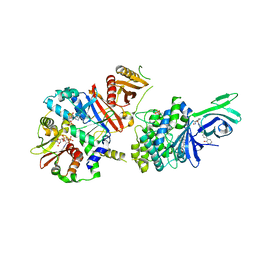 | | AMP-activated protein kinase bound to pharmacological activator R739 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6DK4
 
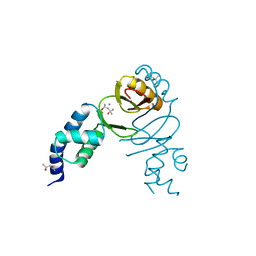 | | Crystal structure of Campylobacter jejuni peroxide stress regulator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ferric uptake regulation protein, MANGANESE (II) ION, ... | | Authors: | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of Campylobacter jejuni peroxide regulator.
FEBS Lett., 592, 2018
|
|
6E2H
 
 | |
6E29
 
 | |
5C6Q
 
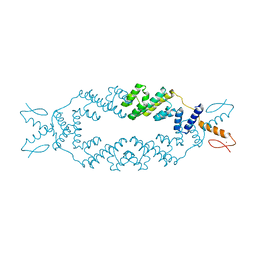 | | Crystal structure of the apo TOPLESS related protein 2 (TPR2) N-terminal domain (1-209) from rice | | Descriptor: | ASPR2 protein, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
5C7E
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal domain (1-209) in complex with Arabidopsis IAA10 peptide | | Descriptor: | ASPR2 protein, Auxin-responsive protein IAA10, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
5C9M
 
 | | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.362 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution.
To Be Published
|
|
5DF5
 
 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
5EQV
 
 | | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-MALATE, FE (III) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site.
To Be Published
|
|
5C7F
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal domain (1-209) in complex with Arabidopsis IAA1 peptide | | Descriptor: | ASPR2 protein, Auxin-responsive protein IAA1, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
5C6V
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal domain (1-209) in complex with Arabidopsis NINJA peptide | | Descriptor: | AFP homolog 2, ASPR2 protein | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
7JZ0
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
2NWF
 
 | | Soluble domain of Rieske Iron Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
