7BJU
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI08346 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
4RU3
 
 | |
4S36
 
 | |
7BJV
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI09181 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
4RU4
 
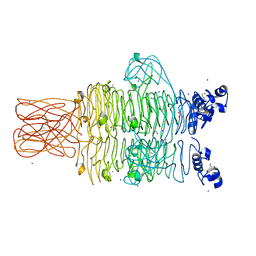 | | Crystal structure of the tailspike protein gp49 from Pseudomonas phage LKA1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Browning, C, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
4RU5
 
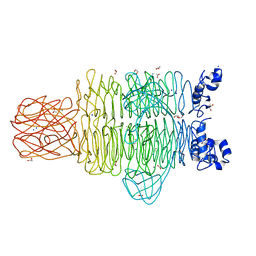 | | Crystal Structure of the Pseudomonas phage phi297 tailspike gp61 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Browning, C, Sycheva, L.V, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
4S37
 
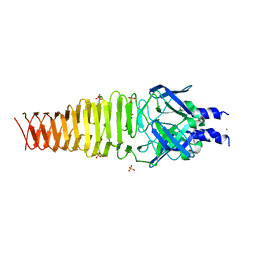 | |
3PQI
 
 | |
3QR8
 
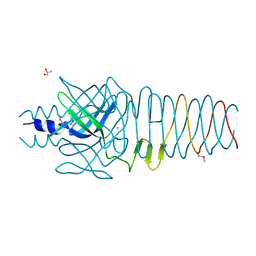 | |
3QR7
 
 | | Crystal structure of the C-terminal fragment of the bacteriophage P2 membrane-piercing protein gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Browning, C, Shneider, M, Leiman, P.G. | | Deposit date: | 2011-02-17 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Phage pierces the host cell membrane with the iron-loaded spike.
Structure, 20, 2012
|
|
3PQH
 
 | |
6ED7
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772 | | Descriptor: | 2-[(2-nitrophenyl)sulfanyl]acetohydrazide, 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Brown, C.M, Zlitni, S, Chan, J, Brown, E.D, Junop, M.S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772
To Be Published
|
|
7XTP
 
 | | eIF4E in Complex with a Disulphide-Free Autonomous VH Domain | | Descriptor: | Eukaryotic translation initiation factor 4E, VH-S4ss, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Brown, C.J, Frosi, Y, Jiang, S, Lin, Y.C. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex.
Nat Commun, 13, 2022
|
|
1QWX
 
 | | Crystal Structure of a Staphylococcal Inhibitor/Chaperone | | Descriptor: | cysteine protease | | Authors: | Brown, C.K, Gu, Z.-Y, Nickerson, N, McGavin, M.J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-02-10 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
6V5E
 
 | | Crystal structure of CTX-M-14 P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V7T
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6BCT
 
 | | I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (27-MER), ... | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BCI
 
 | | Wild-type I-LtrI bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
2V8X
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | 7-BENZYL GUANINE MONOPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
2V8Y
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1, P-FLUORO-7-BENZYL GUANINE MONOPHOSPHATE | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
6BDG
 
 | | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution | | Descriptor: | RNA-binding protein Hfq | | Authors: | Brown, C, Zhang, K, Seo, C, Ellis, M.J, Hanniford, D.B, Junop, M. | | Deposit date: | 2017-10-23 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution
To Be Published
|
|
