4Z58
 
 | | HipB-O3 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
3TPD
 
 | | Structure of pHipA, monoclinic form | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3T03
 
 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
5HJZ
 
 | |
5HK3
 
 | |
5HKC
 
 | |
5HUV
 
 | |
5HVL
 
 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase [UDP-forming], SULFATE ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5HVO
 
 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase B in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
3TPV
 
 | | Structure of pHipA bound to ADP | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T.M, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPT
 
 | | Structure of HipA(D309Q) bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
1Z91
 
 | |
3GIB
 
 | |
1Z9C
 
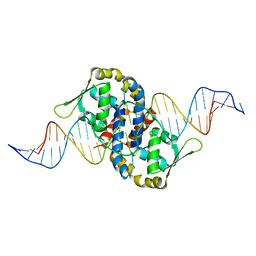 | | Crystal structure of OhrR bound to the ohrA promoter: Structure of MarR family protein with operator DNA | | Descriptor: | DNA (29-MER), Organic hydroperoxide resistance transcriptional regulator | | Authors: | Hong, M, Fuangthong, M, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2005-04-01 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family.
Mol.Cell, 20, 2005
|
|
1Z3E
 
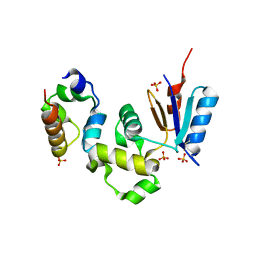 | | Crystal Structure of Spx in Complex with the C-terminal Domain of the RNA Polymerase Alpha Subunit | | Descriptor: | DNA-directed RNA polymerase alpha chain, Regulatory protein spx, SULFATE ION | | Authors: | Newberry, K.J, Nakano, S, Zuber, P, Brennan, R.G. | | Deposit date: | 2005-03-11 | | Release date: | 2005-10-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the {alpha} C-terminal domain of RNA polymerase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZAY
 
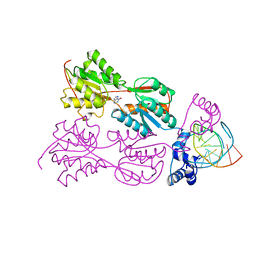 | | PURINE REPRESSOR-HYPOXANTHINE-MODIFIED-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*TP*(1AP)P*TP*TP*TP*TP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Zheleznova, E.E, Brennan, R.G. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Roles of Exocyclic Groups in the Central Base-Pair Step in Modulating the Affinity of PurR for its Operator
To be Published
|
|
1ZVV
 
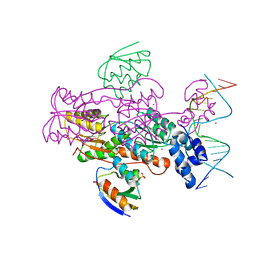 | | Crystal structure of a ccpa-crh-dna complex | | Descriptor: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | Authors: | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | Deposit date: | 2005-06-02 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
2BOW
 
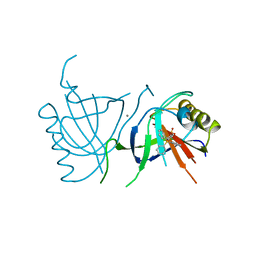 | | MULTIDRUG-BINDING DOMAIN OF TRANSCRIPTION ACTIVATOR BMRR IN COMPLEX WITH A LIGAND, TETRAPHENYLPHOSPHONIUM | | Descriptor: | MANGANESE (II) ION, MULTIDRUG-EFFLUX TRANSPORTER 1 REGULATOR BMRR, SULFATE ION, ... | | Authors: | Zheleznova, E.E, Markham, P.N, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter.
Cell(Cambridge,Mass.), 96, 1999
|
|
3BTI
 
 | |
3BTC
 
 | |
3BTJ
 
 | |
3BTL
 
 | |
3BT9
 
 | |
3D6Z
 
 | |
3D6Y
 
 | |
