7QXV
 
 | | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hemin transport protein | | Authors: | Barker, P.D, Keith, A, Brear, P, Wales, D. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica
To Be Published
|
|
6YJF
 
 | | Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inhibitor from PEGSmear at pH 6.5 | | Descriptor: | (2~{S})-2-(5-nitrofuran-2-yl)-2,3,5,6,7,8-hexahydro-1~{H}-[1]benzothiolo[2,3-d]pyrimidin-4-one, GLYCEROL, Phosphoglycerate kinase | | Authors: | Hyvonen, M, Brear, P, Blaszczyk, B.K. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phosphoglycerate Kinase as a potential target for antimalarial therapy
to be published
|
|
6YV5
 
 | | Crystal Structure of Serine protease SplB N3Q/S154R from Staphylococcus aureus | | Descriptor: | SODIUM ION, Serine protease | | Authors: | Rangel Pereira, M.R, Brear, P, Knyphausen, P, Jermutus, L, Hollfelder, F. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Serine protease SplB N3Q/S154R from Staphylococcus aureus
To Be Published
|
|
6YV6
 
 | | Crystal Structure of Serine protease SplB N2K/N3Q/S154R from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Serine protease | | Authors: | Rangel Pereira, M.R, Brear, P, Knyphausen, P, Jermutus, L, Hollfelder, F. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal Structure of Serine protease SplB N2K/N3Q/S154R from Staphylococcus aureus
to be published
|
|
7QZU
 
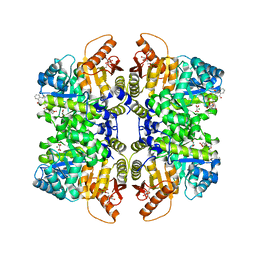 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 47 | | Descriptor: | (2~{S})-2-[2-[4-[3,4-bis(oxidanyl)-9,10-bis(oxidanylidene)anthracen-2-yl]sulfonylpiperazin-1-yl]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2022-01-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
7QUX
 
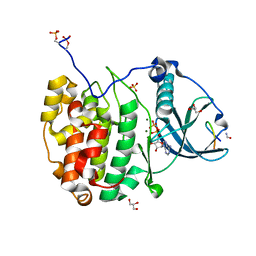 | | Crystal structure of P7C8 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2022-01-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6Z19
 
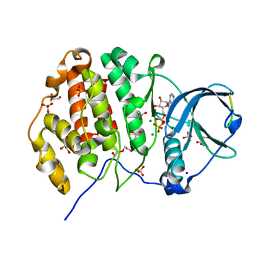 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6S7U
 
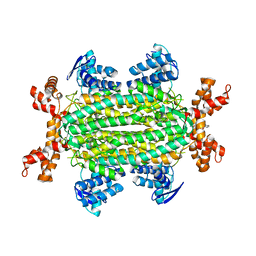 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(1H-indol-3-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S43
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azocan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-06-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S7W
 
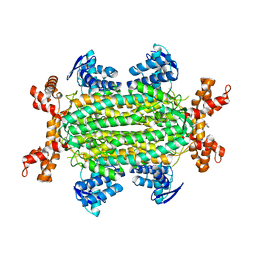 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(quinolin-4-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S7K
 
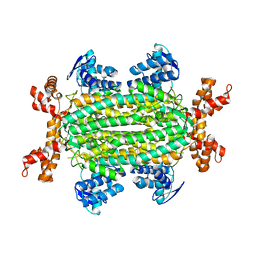 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-(N-methylsulfamoyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S7S
 
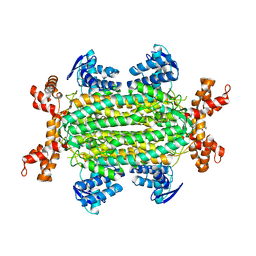 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-(N-phenylsulfamoyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, ~{N}-[2-methoxy-5-(phenylsulfamoyl)phenyl]-2-(4-oxidanylidene-3~{H}-phthalazin-1-yl)ethanamide | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S88
 
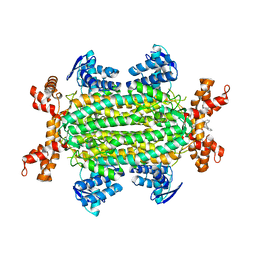 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-((1,2,4,5-tetrahydro-3H-benzo[d]azepin-3-yl)sulfonyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S7Z
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-((3,4-Dihydroisoquinolin-2(1H)-yl)sulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
8BE1
 
 | | SARS-CoV-2 RBD in complex with a Fab fragment of a neutralising antibody mRBD2 | | Descriptor: | Antibody heavy chain, Antibody light chain, SULFATE ION, ... | | Authors: | Lulla, A, Brear, P, Fischer, K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Microfluidics-enabled fluorescence-activated cell sorting of single pathogen-specific antibody secreting cells for the rapid discovery of monoclonal antibodies
Biorxiv, 2023
|
|
6YZH
 
 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6ZQT
 
 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427H/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
6ZRN
 
 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427S/L429M/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
7A03
 
 | | The Structure of CHT | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandes, G, Machado, E, Pereira, M, Brear, P, Lemos, E. | | Deposit date: | 2020-08-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Exploring the genome of Chitinophaga (CB10) for metalocarboxipeptidase activity
To Be Published
|
|
6S6F
 
 | |
6T5M
 
 | |
6ZU0
 
 | |
6S87
 
 | |
6S62
 
 | |
4XYX
 
 | | NanB plus Optactamide | | Descriptor: | Optactamide, PHOSPHATE ION, Sialidase B | | Authors: | Rogers, G.W, Brear, P, Yang, L, Taylor, G.L, Westwood, N.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To Be Published
|
|
