1GGK
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201HIS VARIANT. | | Descriptor: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
5CXC
 
 | | Structure of Ytm1 bound to the C-terminal domain of Erb1 in P 65 2 2 space group | | Descriptor: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | Authors: | Wegrecki, M, Bravo, J. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5CYK
 
 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | Descriptor: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | Authors: | Wegrecki, M, Bravo, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
4A9A
 
 | | Structure of Rbg1 in complex with Tma46 dfrp domain | | Descriptor: | RIBOSOME-INTERACTING GTPASE 1, TRANSLATION MACHINERY-ASSOCIATED PROTEIN 46 | | Authors: | Francis, S.M, Gas, M, Daugeron, M, Seraphin, B, Bravo, J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rbg1-Tma46 Dimer Structure Reveals New Functional Domains and Their Role in Polysome Recruitment.
Nucleic Acids Res., 40, 2012
|
|
5CXB
 
 | |
5D78
 
 | |
1GG9
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ASN VARIANT. | | Descriptor: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
5D77
 
 | | Structure of Mip6 RRM3 Domain | | Descriptor: | CITRIC ACID, NITRATE ION, RNA-binding protein MIP6, ... | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mip6 RRM3 domain at 1.3
To Be Published
|
|
8DFS
 
 | | type I-C Cascade bound to AcrIF2 | | Descriptor: | Anti-CRISPR protein 30, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DFO
 
 | | type I-C Cascade bound to AcrIC4 | | Descriptor: | AcrIC4, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DEJ
 
 | | D. vulgaris type I-C Cascade bound to dsDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DFA
 
 | | type I-C Cascade bound to ssDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DEX
 
 | | type I-C Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
2J6K
 
 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) | | Descriptor: | CD2-ASSOCIATED PROTEIN, SODIUM ION | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J7I
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETERODIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal Src Homology 3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6O
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETEROTRIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2IWL
 
 | |
2J6F
 
 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) BOUND TO CBL-B PEPTIDE | | Descriptor: | CD2-ASSOCIATED PROTEIN, E3 UBIQUITIN-PROTEIN LIGASE CBL-B | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-09-28 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
1E50
 
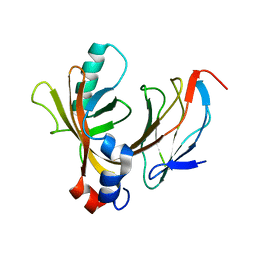 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
1EVS
 
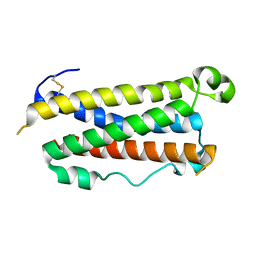 | | CRYSTAL STRUCTURE OF HUMAN ONCOSTATIN M | | Descriptor: | ONCOSTATIN M | | Authors: | Deller, M.C, Hudson, K.R, Ikemizu, S, Bravo, J, Jones, E.Y, Heath, J.K. | | Deposit date: | 2000-04-20 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional dissection of the cytostatic cytokine oncostatin M.
Structure Fold.Des., 8, 2000
|
|
8T7S
 
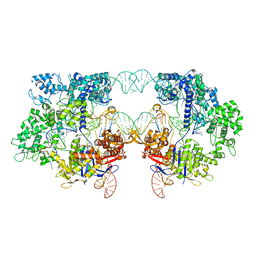 | | SpRYmer bound to NAC PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8SRS
 
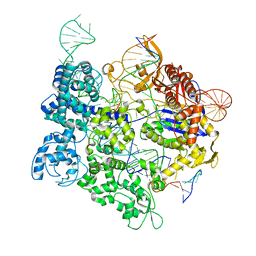 | | SpRY-Cas9:gRNA complex targeting TAC PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-05-06 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8T76
 
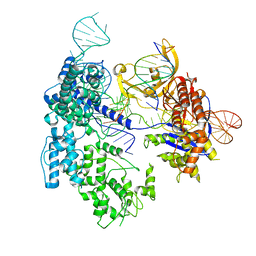 | | SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 3 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8T6P
 
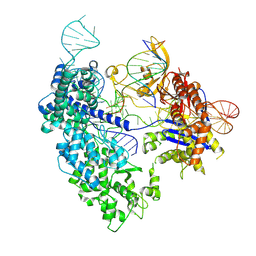 | | SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 2 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-16 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8SPQ
 
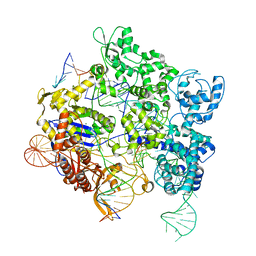 | | SpRY-Cas9:gRNA complex targeting TGG PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
