5IOM
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Schistosoma mansoni is space group P6322 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Torini, J.R.S, Romanello, L, Bird, L.E, Nettleship, J.E, Owens, R.J, Aller, P, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-03-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Schistosoma mansoni NDPK expressed in sexual and digestive organs.
Mol.Biochem.Parasitol., 2019
|
|
5F7X
 
 | | Crystal structure of Mutant Q289L of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in complex with Tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5F7Z
 
 | | Crystal structure of Double Mutant S12T and N87T of Adenosine/Methylthioadenosine Phosphorylase from Schistosoma mansoni in APO Form | | Descriptor: | Methylthioadenosine phosphorylase, PHOSPHATE ION | | Authors: | Torini, J.R, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-08 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5IRR
 
 | | Crystal structure of Septin GTPase domain from Chlamydomonas reinhardtii | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Septin-like protein | | Authors: | Pinto, A.P.A, Pereira, H.M, Navarro, M.V.A.S, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Filaments and fingers: Novel structural aspects of the single septin from Chlamydomonas reinhardtii.
J. Biol. Chem., 292, 2017
|
|
6G5P
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment FM009493b 2,3-Dimethoxy-2,3-dimethyl-2,3-dihydro-1,4-benzodioxin-6-amine | | Descriptor: | (2~{R},3~{R})-2,3-dimethoxy-2,3-dimethyl-1,4-benzodioxin-6-amine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
6G5N
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment XS039818e 1-(3-Phenyl-1,2,4-oxadiazol-5-yl)methanamine | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
5D9G
 
 | | Crystal structure of TIPRL, TOR signaling pathway regulator-like, in complex with peptide | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Scorsato, V, Sandy, J, Brandao-Neto, J, Pereira, H.M, Smetana, J.H.C, Aparicio, R. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human Tip41 orthologue, TIPRL, reveals a novel fold and a binding site for the PP2Ac C-terminus.
Sci Rep, 6, 2016
|
|
5CXS
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Purine nucleoside phosphorylase | | Authors: | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
5EYT
 
 | | Crystal Structure of Adenylosuccinate Lyase from Schistosoma mansoni in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L, Nettleship, J, Owens, R, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3649 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
5F78
 
 | | Crystal structure of Mutant N87T of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8518 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5F73
 
 | | Crystal structure of Mutant S12T of Adenosine/Methylthioadenosine Phosphorylase in APO form | | Descriptor: | Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5F77
 
 | | Crystal structure of Mutant S12T of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in complex with Adenine | | Descriptor: | ADENINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5CXQ
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | Purine nucleoside phosphorylase | | Authors: | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
5F76
 
 | | Crystal structure of Mutant S12T of Adenosine/Methylthioadenosine Phosphorylase from Schistosoma mansoni in complex with Methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5EYV
 
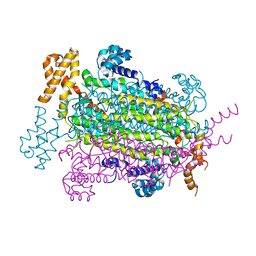 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
5AMF
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment Ethyl 4,5,6,7- tetrahydro-1H-indazole-5-carboxylate (SGC - Diamond I04-1 fragment screening) | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 1, ETHYL (5R)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE-5-CARBOXYLATE, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment Ethyl 4,5,6,7-Tetrahydro-1H-Indazole-5-Carboxylate
To be Published
|
|
5AME
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment 4-acetyl- piperazin-2-one (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 4-acetyl-piperazin-2-one, BROMODOMAIN-CONTAINING PROTEIN 1, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment 4-Acetyl-Piperazin-2-One
To be Published
|
|
5B8D
 
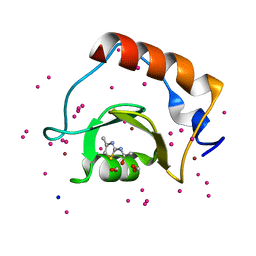 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | Authors: | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
7NW2
 
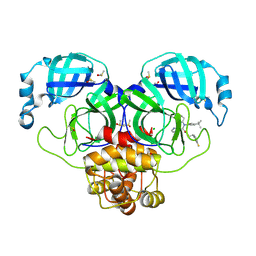 | | Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-47 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Douangamath, A, Aimon, A, Brandao-Neto, J, Dias, A, Dunnett, L, Gehrtz, P, Gorrie-Stone, T.J, Lukacik, P, Powell, A.J, Skyner, R, Strain-Damerell, C.M, Zaidman, D, London, N, Walsh, M.A, von Delft, F, Covid Moonshot Consortium | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 M pro inhibitor.
Cell Chem Biol, 28, 2021
|
|
5KCH
 
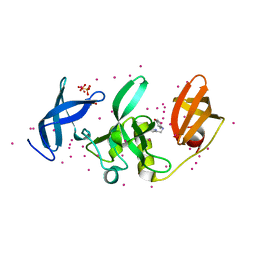 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy into weak electron density | | Descriptor: | 4-methoxy-N-[(pyridin-2-yl)methyl]aniline, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5KCO
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy | | Descriptor: | DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5KH9
 
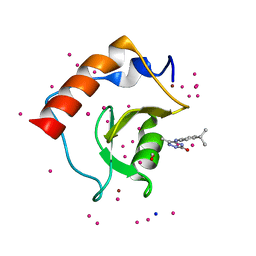 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5L16
 
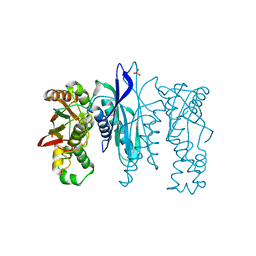 | | Crystal Structure of N-terminus truncated selenophosphate synthetase from Leishmania major | | Descriptor: | Putative selenophosphate synthetase, SULFATE ION | | Authors: | Faim, L.M, Silva, I.R, Pereira, H.M, Dias, M.B, Silva, M.T.A, Brandao-Neto, J, Thiemann, O.H. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Trypanosomatid selenophosphate synthetase structure, function and interaction with selenocysteine lyase.
Plos Negl Trop Dis, 14, 2020
|
|
6ZGZ
 
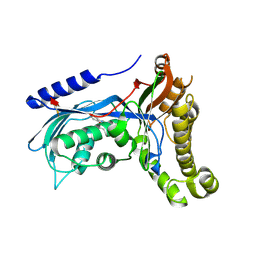 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6ZGX
 
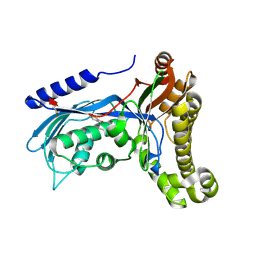 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
