8R5K
 
 | |
1A8R
 
 | | GTP CYCLOHYDROLASE I (H112S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-27 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I.
J.Mol.Biol., 326, 2003
|
|
1A9C
 
 | | GTP CYCLOHYDROLASE I (C110S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-04-04 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GTP Cyclohydrolase I in Complex with GTP at 2.1 A Resolution
To be Published
|
|
2PEN
 
 | | Crystal structure of RbcX, crystal form I | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEJ
 
 | | Crystal structure of RbcX point mutant Y17A/Y20L | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEO
 
 | | Crystal structure of RbcX from Anabaena CA | | Descriptor: | RbcX protein | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PEK
 
 | | Crystal structure of RbcX point mutant Q29A | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
6Z1G
 
 | | CryoEM structure of the interaction between Rubisco Activase small-subunit-like (SSUL) domain with Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, Ribulose bisphosphate carboxylase/oxygenase activase | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
6ZCO
 
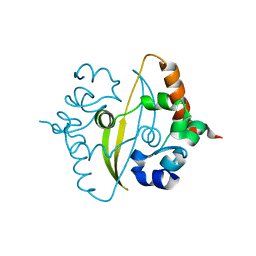 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | Descriptor: | Nucleoprotein | | Authors: | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
2WVW
 
 | | Cryo-EM structure of the RbcL-RbcX complex | | Descriptor: | RBCX PROTEIN, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN | | Authors: | Liu, C, Young, A.L, Starling-Windhof, A, Bracher, A, Saschenbrecker, S, Rao, B.V, Rao, K.V, Berninghausen, O, Mielke, T, Hartl, F.U, Beckmann, R, Hayer-Hartl, M. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Coupled Chaperone Action in Folding and Assembly of Hexadecameric Rubisco
Nature, 463, 2010
|
|
8QXS
 
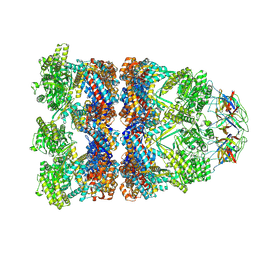 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with wide GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXT
 
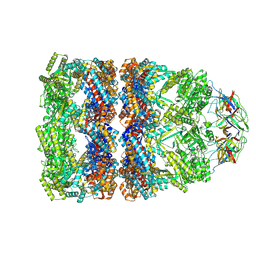 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with narrow GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXU
 
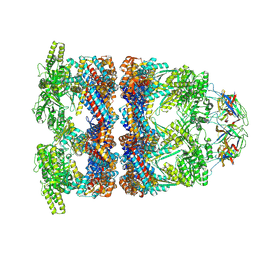 | | In situ structure average of GroEL14-GroES7 complexes with wide GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXV
 
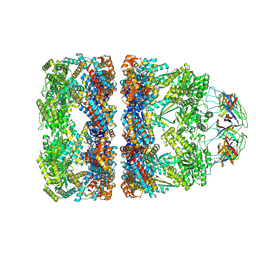 | | In situ structure average of GroEL14-GroES7 complexes with narrow GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
7AOT
 
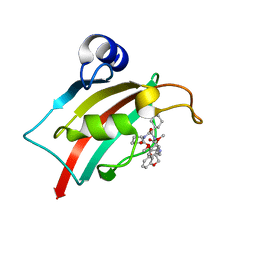 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5OBK
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Sippel, C, Haehle, A, Bracher, A, Hausch, F. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chemogenomic Profiling of Human and Microbial FK506-Binding Proteins.
J. Med. Chem., 61, 2018
|
|
4TW8
 
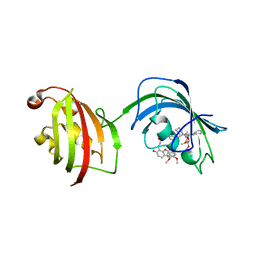 | | The Fk1-Fk2 domains of FKBP52 in complex with iFit-FL | | Descriptor: | 2-(5-{[({3-[(1R)-1-[({(2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetyl)amino]methyl}-6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
1XQR
 
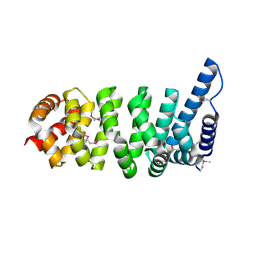 | | Crystal structure of the HspBP1 core domain | | Descriptor: | HspBP1 protein | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
2AZ1
 
 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum | | Descriptor: | CALCIUM ION, Nucleoside diphosphate kinase | | Authors: | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | Deposit date: | 2005-09-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
1XQS
 
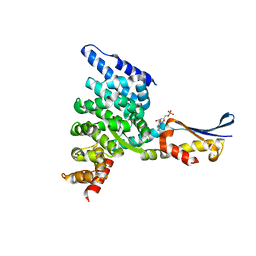 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
2AZ3
 
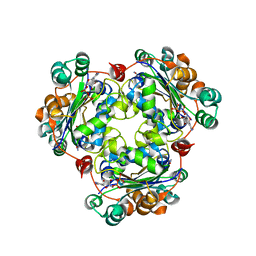 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum in complex with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | Deposit date: | 2005-09-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
4TW6
 
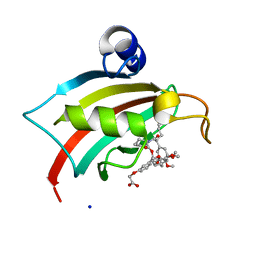 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
4TW7
 
 | | The Fk1 domain of FKBP51 in complex with iFit4 | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
4WT3
 
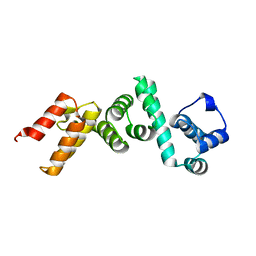 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT4
 
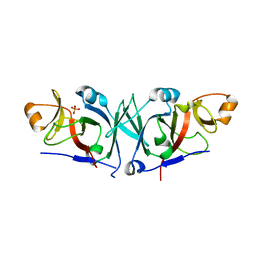 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
