2ASR
 
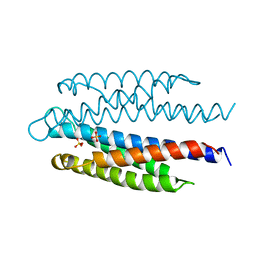 | |
1F0M
 
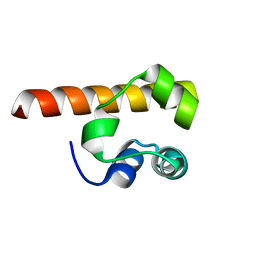 | | MONOMERIC STRUCTURE OF THE HUMAN EPHB2 SAM (STERILE ALPHA MOTIF) DOMAIN | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Thanos, C.D, Faham, S, Goodwill, K.E, Cascio, D, Phillips, M, Bowie, J.U. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monomeric structure of the human EphB2 sterile alpha motif domain.
J.Biol.Chem., 274, 1999
|
|
2QB0
 
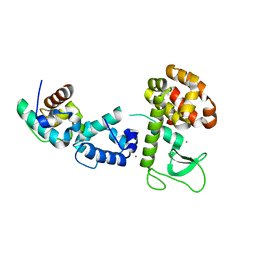 | |
6VGS
 
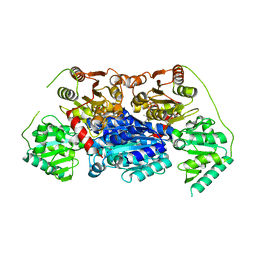 | | Alpha-ketoisovalerate decarboxylase (KivD) from Lactococcus lactis, thermostable mutant | | Descriptor: | Alpha-keto acid decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Chan, S, Korman, T.P, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2020-01-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isobutanol production freed from biological limits using synthetic biochemistry.
Nat Commun, 11, 2020
|
|
7T71
 
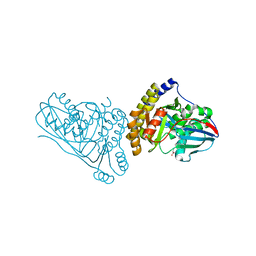 | | Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus | | Descriptor: | Mevalonate 3,5-bisphosphate decarboxylase, OLEIC ACID | | Authors: | Vinokur, J.M, Sawaya, M.R, Cascio, D, Collazo, M, Bowie, J.U. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways.
J.Biol.Chem., 298, 2022
|
|
3BQ7
 
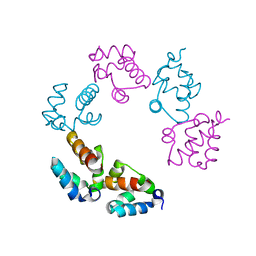 | |
1B4F
 
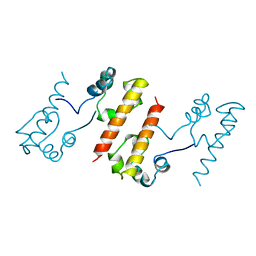 | |
1XJI
 
 | | Bacteriorhodopsin crystallized in bicelles at room temperature | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Faham, S, Boulting, G.L, Massey, E.A, Yohannan, S, Yang, D, Bowie, J.U. | | Deposit date: | 2004-09-23 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization of bacteriorhodopsin from bicelle formulations at room temperature
Protein Sci., 14, 2005
|
|
2QAR
 
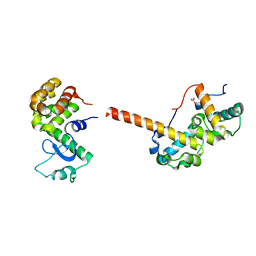 | |
2QB1
 
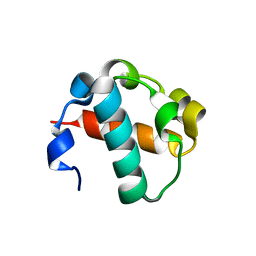 | | 2TEL crystallization module | | Descriptor: | E80-TELSAM domain | | Authors: | Nauli, S, Bowie, J.U. | | Deposit date: | 2007-06-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Polymer-driven crystallization.
Protein Sci., 16, 2007
|
|
3COC
 
 | |
3COD
 
 | | Crystal Structure of T90A/D115A mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Joh, N.H, Min, A, Faham, S, Bowie, J.U. | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins.
Nature, 453, 2008
|
|
4NJ8
 
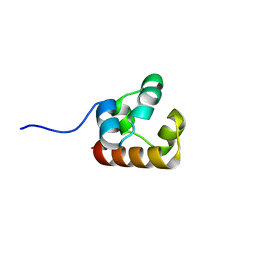 | |
4NL9
 
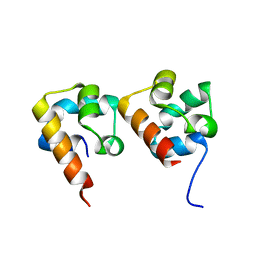 | |
2F3N
 
 | |
2F44
 
 | |
4GXN
 
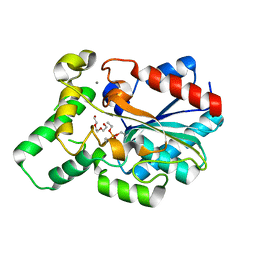 | |
1KME
 
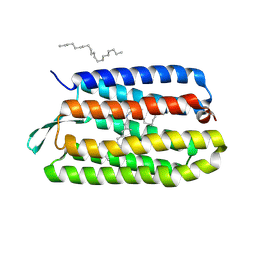 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN CRYSTALLIZED FROM BICELLES | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, RETINAL, ... | | Authors: | Faham, S, Bowie, J.U. | | Deposit date: | 2001-12-14 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure.
J.Mol.Biol., 316, 2002
|
|
1KW4
 
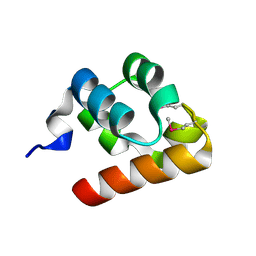 | |
1JI7
 
 | | Crystal Structure of TEL SAM Polymer | | Descriptor: | ETS-RELATED PROTEIN TEL1, SULFATE ION | | Authors: | Kim, C.A, Phillips, M.L, Kim, W, Gingery, M, Tran, H.H, Robinson, M.A, Faham, S, Bowie, J.U. | | Deposit date: | 2001-06-29 | | Release date: | 2002-07-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression.
EMBO J., 20, 2001
|
|
4RKP
 
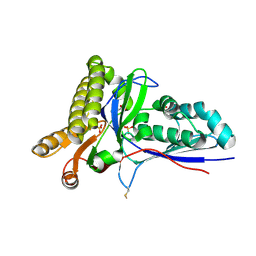 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (apo form) | | Descriptor: | ACETATE ION, Putative uncharacterized protein Ta1305, SULFATE ION | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RKZ
 
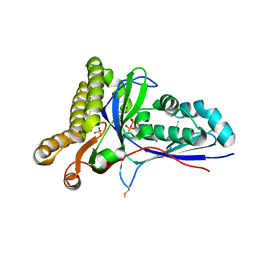 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate 3-Phosphate/ADP Bound) | | Descriptor: | (3R)-5-hydroxy-3-methyl-3-(phosphonooxy)pentanoic acid, ADENOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein Ta1305, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RKS
 
 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate Bound) | | Descriptor: | (R)-MEVALONATE, ACETATE ION, GLYCEROL, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RQN
 
 | | Crystal structure of the native BICC1 SAM Domain R924E mutant | | Descriptor: | Protein bicaudal C homolog 1, ZINC ION | | Authors: | Leettola, C.N, Cascio, D, Bowie, J.U. | | Deposit date: | 2014-11-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bicc1 SAM Polymer and Mapping of Interactions between the Ciliopathy-Associated Proteins Bicc1, ANKS3, and ANKS6.
Structure, 26, 2018
|
|
4RQM
 
 | | Crystal structure of the SeMET BICC1 SAM Domain R924E mutant | | Descriptor: | Protein bicaudal C homolog 1, ZINC ION | | Authors: | Leettola, C.N, Cascio, D, Bowie, J.U. | | Deposit date: | 2014-11-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Bicc1 SAM Polymer and Mapping of Interactions between the Ciliopathy-Associated Proteins Bicc1, ANKS3, and ANKS6.
Structure, 26, 2018
|
|
